ABOVE: © istock.com, luismmolin, Design Cells
Humans are never alone. Even in a room devoid of other people, they are always in the company of billions of microscopic beings. By residing in and on the human body, these bacteria, fungi, and viruses form complex ecosystems that exert powerful effects on a range of health conditions, such as antitumor responses, gut inflammatory diseases, and mental health.1-3
With such striking effects, many scientists see modulating the microbiomes as a promising avenue for improving human health and wellbeing. In recent years, researchers have shown that tweaking the genomes of resident microorganisms offers new ways to treat and diagnose diseases.4-6 Despite these advances, most efforts to engineer microbes focus on a handful of well characterized and genetically tractable microbes, leaving most of these microbial communities in uncharted territories.
We can target harmful bacteria. We can engineer beneficial bacteria. We can use CRISPR to study microbial interactions. We can use CRISPR to remove antibiotic resistance genes. It could be quite powerful in a microbiome context.
—James Marsh, Max Planck Institute for Biology
When looking at widely studied microbiomes such as the human gut microbiome, less than 10 percent of the constituent microbes can be genetically manipulated, according to James Marsh, a microbiome engineer at the Max Planck Institute for Biology. “They’re usually what we call model organisms because we’ve been studying them for decades, like E. coli. And that’s where we run into a problem: When we try to apply traditional techniques that have been established for E. coli, they don’t apply to the microbiome,” said Marsh. One primary reason is that different microbial species have varied requirements for uptake of foreign DNA. Scientists also face challenges optimizing the culture conditions when isolating unknown microbiome inhabitants.
“We really need to work out a way to access the majority rather than the model organism minority,” said Marsh.
In their quest for ideal molecular tools that can be easily adapted from one species to another, many scientists are turning their attention to CRISPR. This genome-editing tool allows researchers to precisely target one or more genes simultaneously in the same organism, and the ease with which the technology can be adapted from one organism to another makes it ideal for genetically manipulating a multitude of microbial species.
CRISPR: from prokaryotes to eukaryotes and back
In nature, CRISPR, short for clustered regularly interspaced short palindromic repeats, is an adaptive immune system in prokaryotes, namely bacteria and archaea.7,8 When a virus infects a bacterium, for instance, the latter stores a snippet of the viral DNA as spacers between repeats in its own genome to retain a memory of the infection. If the same virus invades again, the bacterium transcribes guide RNA (gRNA) from these viral fragments, which attaches to the viral DNA and tags it for cleavage by CRISPR-associated (Cas) proteins.

Scientists have categorized different types of CRISPR systems into two classes based on how their Cas nucleases function. In class 1 (types I, III, and IV), different Cas proteins form a complex machinery to identify and cut foreign DNA; in class 2 CRISPR systems (types II, V, and VI), a single Cas protein effector recognizes and cleaves DNA.9
After characterizing CRISPR’s role as a defense mechanism in bacteria, researchers soon realized that they could harness this system for gene manipulation in any cell. All they needed to do was design a CRISPR gRNA sequence that bound to a specific DNA sequence and triggered the Cas nuclease, which would then cut precisely at that location. With CRISPR, researchers routinely knock out gene function by cutting out a DNA fragment, or they insert a desired genetic sequence into the genome by providing a reference DNA template along with the CRISPR components. While editing eukaryotic cells has been the focus for tackling diseases, many researchers now use CRISPR to edit bacterial communities.
“It’s almost like back to the beginning or back to the origins. There’s some irony in bringing CRISPR back to where it came from,” said Rodolphe Barrangou, a functional genomics researcher at North Carolina State University, who helped characterize the immune function of CRISPR and has been working with it for more than 20 years.
CRISPR-based technologies can be quite powerful in the journey to genetically manipulate the microbiome, according to Marsh. “We can target harmful bacteria. We can engineer beneficial bacteria. We can use CRISPR to study microbial interactions. We can use CRISPR to remove antibiotic resistance genes. It could be quite powerful in a microbiome context,” said Marsh.
Repurposing native CRISPR systems
To engineer microbes within the microbiome, scientists can leverage an existing CRISPR system, an approach that Barrangou has been exploring for more than a decade. “Almost half of all bacteria and the large majority of archaea on planet Earth have an endogenous CRISPR-Cas system in them,” he said. “One of the great things to do is just to repurpose the endogenous machinery.”
As intuitive as this idea may sound, exploiting endogenous CRISPR systems can be cumbersome; scientists must first figure out if they work and then test if they can be reused. But there are also advantages to this approach. For instance, researchers avoid the potential cytotoxicity exogenous Cas effectors might have. Additionally, as plasmids do not need to pack entire CRISPR systems, they are smaller in size, which might favor their delivery.

In his lab, Barrangou explores how to harness a widespread endogenous type I CRISPR system and its associated nuclease, Cas3, to potentially enhance the efficacy of probiotic bacteria such as Bifidobacterium and Lactobacillus. Bifidobacterium species are typical gut bacteria, and their presence associates with many health benefits, including improving host immunity and preventing gut infections.10 Lactobacilli also inhabit the human gut, and some species improve intestinal barrier function in mice.11
In a series of in vitro studies in Bifidobacterium lactis and Lactobacillus crispatus, Barrangou’s team repurposed the type I CRISPR-Cas systems that they harbor as effective gene-editing tools. The researchers showed that both CRISPR systems from both bacteria could be repurposed for genome editing by delivering a plasmid containing a self-targeting gRNA that directed the multi-subunit cascade complex and the Cas3 nuclease to the target genomic site and repair templates that enabled gene deletion without lethal consequences for the bacteria.12,13
Beyond improving the efficacy of probiotics, Barrangou was also interested in reprogramming endogenous CRISPR systems to act as antimicrobial agents. In contrast to broad-spectrum antibiotics that indiscriminately eliminate both pathogenic and beneficial bacteria, CRISPR-based antimicrobials may provide a more precise way to kill pathogenic bacteria without affecting the rest of the microbiota.
Clostridioides difficile (C. difficile) is one such harmful bacterial species that could be targeted by CRISPR antimicrobials. Infections caused by C. difficile affect approximately half a million patients in the United States, leading to 30,000 annual deaths.14 Broad-spectrum antibiotics are commonly used to treat C. difficile illness; however, antibiotic treatment often eliminates commensal bacterial species that help keep C. difficile growth in check in the microbiota.
Barrangou teamed up with Casey Theriot, a microbiologist at North Carolina State University, and David Ousterout, a biomedical engineer at the precision medicine company Locus Biosciences, to use CRISPR self-targeting in C. difficile. Instead of using plasmids to deliver the RNA guide into cells, the team turned their attention to bacteriophages, bacterial viruses that specifically target their bona fide host. “It makes a lot of sense to use phages for very accurate, very precise, narrow spectrum, antimicrobial applications,” said Barrangou.
Using a recombinant phage expressing the CRISPR gRNA targeting a specific DNA sequence, the researchers tested whether this method would redirect the C. difficile CRISPR system to target bacterial DNA in a mouse model of C. difficile infection. Treating mice with phages encoding the CRISPR targeting of C. difficile DNA reduced bacterial burden. Further refinement of the phage delivery vehicles resulted in greater reductions in the inflammation and epithelial damage caused by C. difficile infection, highlighting how phages can be coupled with CRISPR-based targeting to create novel therapies against pathogenic microbes.15
Engineering Microbiomes with CRISPRThe microbes that make up an organism’s microbiome have a range of effects on its health. Scientists use CRISPR systems to genetically manipulate specific bacterial species, for instance those found in the mouse gut microbiome, to find new ways to promote health and treat disease. |
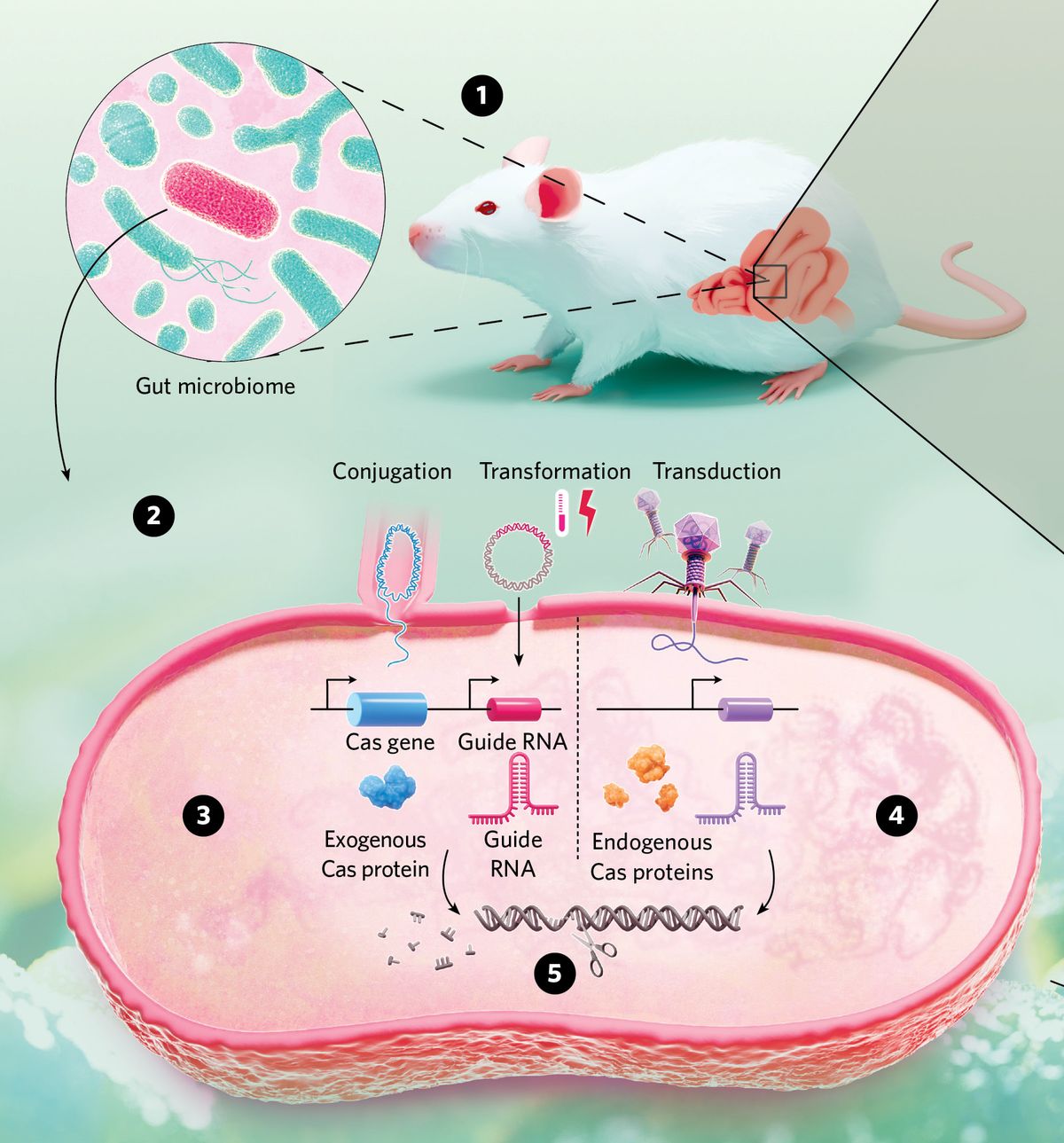 © nanoclustering.com (1) Scientists select a specific microbe, such as a bacterial species, for CRISPR-mediated genetic manipulation. (2) Researchers deliver CRISPR system components using different methods, including conjugation, transformation by heat shock or electroporation, and transduction by bacteriophages. (3) Scientists directly provide the sequence encoding all CRISPR components, a Cas protein and a guide RNA, into the target microbe. (4) Alternatively, scientists co-opt the bacteria’s endogenous CRISPR machinery and supply only the guide RNA. (5) Scientists edit the microbe’s DNA or cause irreparable breaks that lead to DNA degradation. |
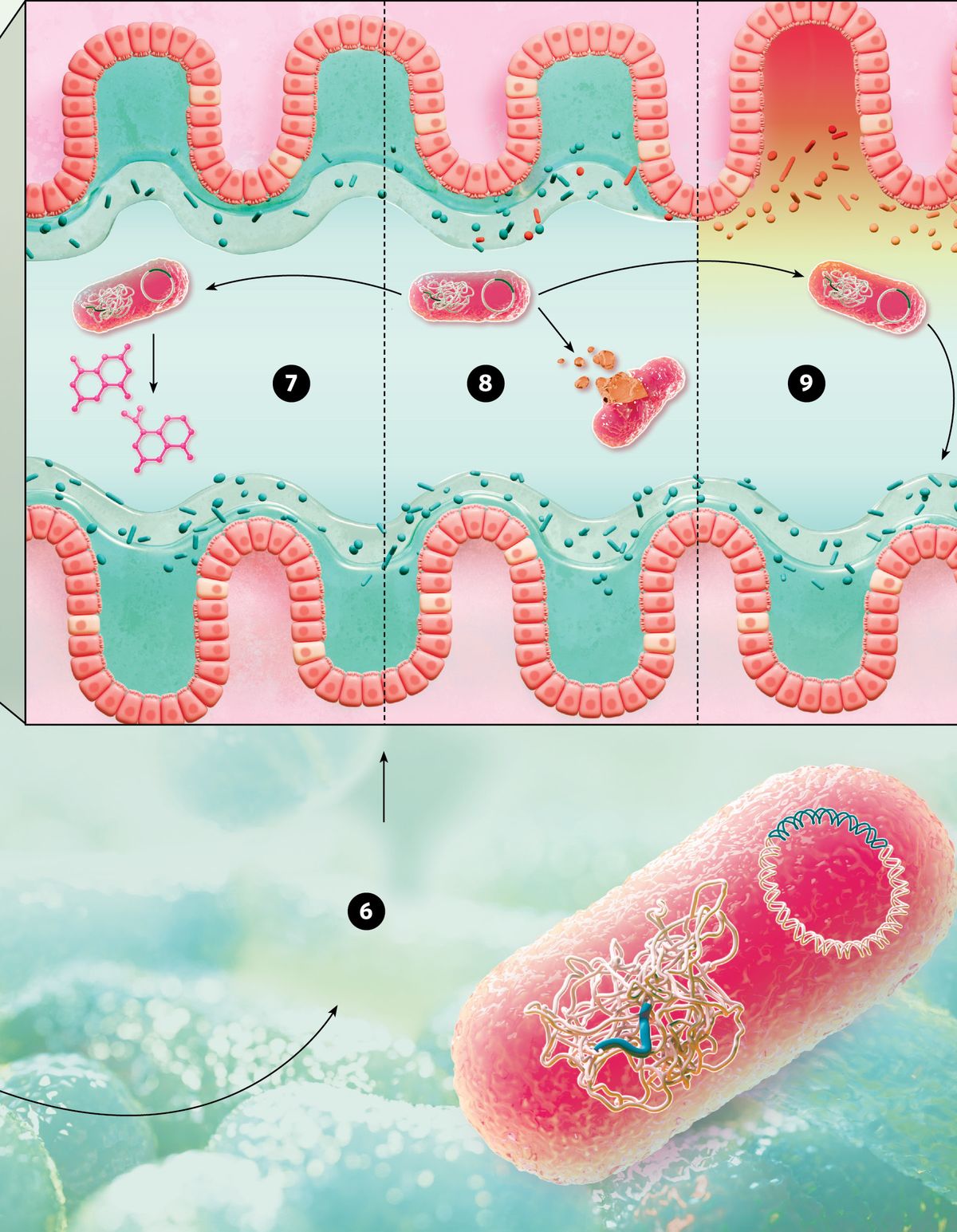 © nanoclustering.com (6) In bacteria, scientists modify either the plasmid or chromosomal DNA of the microbe. In complex microbial communities such as the gut microbiome, CRISPR-engineered bacteria have multiple applications. (7) Engineered probiotic bacteria produce modulatory molecules to fight diseases and restore the intestinal flora. (8) As an antimicrobial, CRISPR self-targets the DNA of a pathogenic bacterial strain. (9) By CRISPR editing commensal gut bacteria, scientists modulate the effects of these microbes on the microbiome and influence disease related processes such as inflammation. |
Bringing in external CRISPR systems
Besides harnessing endogenous CRISPR-Cas systems to engineer human-associated microbes, scientists can also deliver exogenous CRISPR systems to them. For these systems to work, the CRISPR payloads must pack a sequence encoding a gRNA for precise DNA targeting, instructions for one or more Cas effectors for making the genome breaks, and repair templates in case researchers want to introduce specific mutations.
That was attractive from the standpoint of wanting not just to kill the microbe, but even try to actually remove specific genes from the genome [and] see which genes are important for a given phenotype.
—Peter Turnbaugh, University of California, San Francisco
In his lab at the University of California, San Francisco, microbial geneticist Peter Turnbaugh developed an approach that could precisely edit bacterial strains or species in established microbiomes using CRISPR. Turnbaugh’s interest in studying microbial communities dates back to graduate school, when, under the mentorship of Washington University physician scientist Jeffrey Gordon, he explored the relationship between diet and the gut microbiome and how differences in this microbial community contributed to metabolic diseases such as obesity. “That got me hooked, and I’ve been able to keep working on it ever since,” Turnbaugh said.
To manipulate the gut microbiome of a mouse at the species level, Turnbaugh and his team chose the CRISPR-Cas9 system, a type II CRISPR system that uses the endonuclease Cas9 to both recognize and cut the DNA.16 “That was attractive from the standpoint of wanting not just to kill the microbe, but even try to actually remove specific genes from the genome [and] see which genes are important for a given phenotype,” Turnbaugh said.
The team used bacteriophages to deliver the CRISPR package to the targeted microbe in the mouse gut microbiome. As they were venturing into uncharted waters, they decided to work with a well characterized phage-bacteria pair, the M13 phage and E. coli, as their test system. To assess the efficacy of the phage-based CRISPR delivery approach, they colonized mice with E.coli strains expressing green fluorescent protein (GFP), the targeted bacterial locus, or the red-fluorescent protein mCherry as a control before administering the phage treatment to the animals, and analyzed the presence of fluorescent bacteria in mouse stool samples. Phages carrying CRISPR delivered the payload to the targeted bacterial population, eliminating most GFP-containing E. coli in the mouse gut.17
Even though CRISPR systems like the one used by Turnbaugh’s team showed promising results for microbiome editing, a major bottleneck for scientists attempting to manipulate the genomes of gut bacteria was not knowing whether most of the gut inhabitants would be amenable to genetic editing.
CRISPR editing genetically tractable microbes
Finding ways to identify genetically tractable species within a microbial community that could be genetically engineered in their native habitats is a powerful drive for scientists like Brady Cress, a chemical and biological engineer at the Innovative Genomics Institute at the University of California, Berkeley. Cress’ previous experience in developing CRISPR technologies motivated him to join the lab of the CRISPR pioneer Jennifer Doudna at the University of California, Berkeley, where he could continue learning and developing valuable CRISPR technologies. “[CRISPR] is the most powerful technology for engineering biology,” he said.

As a postdoctoral researcher in Doudna’s lab, Cress teamed up with Benjamin Rubin and Spencer Diamond, who were postdoctoral researchers in the labs of Doudna and Jillian Banfield at the time, to devise an approach for identifying and editing the genomes of microbes within a community.18
To survey the genetic editability of a microbial community, Rubin and Diamond developed a sequencing method dubbed Environmental Transformation Sequencing (ET-seq). In this method, scientists expose the microbial community to a randomly integrating DNA editor, a mariner transposase, and subsequently sequence it looking for insertion events and the abundance of each member of the community in a sample.
Researchers obtain a measure of genetic accessibility through bioinformatics quantification of the insertion events normalized to an internal standard. By creating a nine-member synthetic microbial community in which the team added a known amount of an edited Klebsiella michiganensis, the researchers detected insertion events even in bacterial cells that represented less than 0.01 percent of the total population, highlighting the approach’s potential to screen for bacteria amenable to genome editing without the need to cultivate and test individual species.
While the ET-seq approach allowed the team to identify microbes willing to take up exogenous DNA, they still needed a more precise strategy to genetically edit the identified amenable bacteria. Cress and Rubin came up with the idea of using CRISPR-associated transposases to do the job. “You can think of [them] as a really effective cut and paste version of classic CRISPR tools, which we thought would be really useful for making species- and site-specific edits in microbial communities,” Cress explained.
Previous studies from biochemist Samuel Sternberg at Columbia University and molecular biologist Feng Zhang at the Massachusetts Institute of Technology characterized these CRISPR-associated transposases systems for the first time.19,20 However, these systems were not adapted to be delivered into a microbial community for DNA editing. Cress and his colleagues tweaked these previously characterized systems to build a programmable DNA-editing all-in-one RNA-guided CRISPR-Cas transposase (DART) system that was compatible with the ET-seq method. “People can really envision a dart going into the community and precisely hitting a single locus and a single species’ genome,” Cress said.
If we really hope to change the function of what these microbiomes can do, we really need to be manipulating that blueprint, and CRISPR is the tool that allows us to do that very precisely and very efficiently.
—Brady Cress, University of California, Berkeley
To test the efficacy of the DART system to edit DNA, they designed gRNA against specific regions of the K. michiganensis genome and used conjugation to deliver the DART system to the synthetic soil microbial community. Barcodes attached to the transposases in DART allowed the researchers to track the insertions at the community level. In a series of proof-of-concept experiments, they showed that DART can be used to produce gain-of-function and loss-of-function in the targeted species by designing different genetic payloads in the DART vector.
The ET-seq/DART approach was also successful in a more ecologically relevant microbial community, in this case, an infant gut microbiome. By cultivating an infant stool sample ex vivo, the researchers identified E. coli strains that were amenable to genetic editing. Genetic manipulation of some of these strains enriched their presence in the sample, demonstrating the utility of the approach for enriching low-abundance or difficult-to-isolate microbes.
By looking at microbiomes as collections of genes, the ET-seq/DART approach could be a useful tool for targeting disease-associated genes, according to Cress. “If we think about bad genes instead of bad bacteria, we could just direct DART to go in and break that bad gene, leaving the community composition completely intact,” he said. In addition, because the DART system was delivered by conjugation, a naturally occurring process in which a bacterium produces a needle-like structure to inject DNA into neighboring bacterial cells, the researchers could adapt the tool to edit multiple species at once by designing multiple gRNA. “Let’s say there’s a bad gene that exists not just in one species, but in three different species. We should be able to break them all in a single experiment,” Cress noted.
The future of CRISPR microbiomes
Although scientists have a rich history of genetically manipulating isolated microbes in the laboratory, the field of microbiome engineering is still in its infancy, and scientists in it still have many challenges ahead. DNA delivery is one of the biggest technical bottlenecks, according to Barrangou. “If you go into noncanonical organisms—outside of Bacillus, outside E. coli—organisms in which the toolbox is well established, the biggest factor is not CRISPR. The biggest factor is delivery,” he said.
Cress agreed, adding that researchers are working on ways to improve DNA delivery. Some of the strategies they explore include plasmids and the natural mechanisms used to transfer them. “What we really want to do is take advantage of these natural horizontal gene transfer mechanisms [and] just hitch our DNA editors to these plasmids that are naturally able to move around in the community,” Cress explained.

Although researchers are developing novel approaches for genetically engineering microbes in their communities, the tools available to them are still limited, especially when compared to those available for genetics in isolated microbes. “Now, we need to focus on different ways of studying the community and also different ways of targeting organisms, either within a mixed community or by targeting groups of organisms,” Marsh said. “There’s sort of a dual priority to develop these core strategies that are going to be really helpful for humanity, but also to make them accessible to the broader microorganisms [and] develop tools for that,” he added.
CRISPR-based approaches can help expand the genetic toolkit microbiome engineers have available, bringing them a step closer to understanding the roles and interactions between microbial community members and how these shape their communities and influence the organisms’ hosting them. “Every single function that a community of microbes outputs is encoded within their DNA. So, the DNA is really the blueprint that tells us what these microbes can do within that microbiome,” Cress said. “If we really hope to change the function of what these microbiomes can do, we really need to be manipulating that blueprint, and CRISPR is the tool that allows us to do that very precisely and very efficiently.”
Rodolphe Barrangou is cofounder of Intellia Therapeutics and Ancilia Biosciences and scientific advisor to Inari and Invaio Sciences.
- Zhao LY, et al. Role of the gut microbiota in anticancer therapy: from molecular mechanisms to clinical applications.Signal Transduct Target Ther. 2023;8(1):201.
- Imhann F, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67(1):108-119.
- Valles-Colomer M, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 2019;4(4):623–632.
- Bai X, et al. Engineering the gut microbiome. Nat Rev Bioeng. 2023;1(9):665-679.
- Isabella VM, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol. 2018;36(9):857-864.
- Cooper RM, et al. Engineered bacteria detect tumor DNA. Science. 2023;381(6658):682-686.
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526(7571):55-61.
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709-1712.
- Li Y, Peng N. Endogenous CRISPR-Cas system-based genome editing and antimicrobials: review and prospects. Front Microbiol. 2019;10:2471.
- Wong CB, et al. Insights into the reason of Human-Residential Bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol Rev. 2020;44(3):369-385.
- Martín R, et al. The potential probiotic Lactobacillus rhamnosus CNCM I-3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response. Sci Rep. 2019;9(1).
- Pan M, et al. Genomic and epigenetic landscapes drive CRISPR-based genome editing in Bifidobacterium. Proc Natl Acad Sci U S A. 2022;119(30):e2205068119.
- Hidalgo-Cantabrana C, et al. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc Natl Acad Sci U S A. 2019;116(32):15774-15783.
- Feuerstadt P, et al. The burden of CDI in the United States: a multifactorial challenge. BMC Infect Dis. 2023;23(1):132.
- Selle K, et al. In vivo targeting of Clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials. mBio. 2020;11(2):e00019-20.
- Sternberg SH, Doudna JA. Expanding the biologist’s toolkit with CRISPR-Cas9. Mol Cell. 2015;58(4):568-574.
- Lam KN, et al. Phage-delivered CRISPR-Cas9 for strain-specific depletion and genomic deletions in the gut microbiome. Cell Rep. 2021;37(5):109930.
- Rubin BE, et al. Species- and site-specific genome editing in complex bacterial communities. Nat Microbiol. 2022;7(1):34-47.
- Klompe SE, et al. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571(7764):219-225.
- Strecker J, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48-53.
Membership Open House!
Enjoy OPEN access to Premium Content for a limited timeInterested in exclusive access to more premium content?