Stay up to date on the latest science with Brush Up Summaries.
What Is PCR?
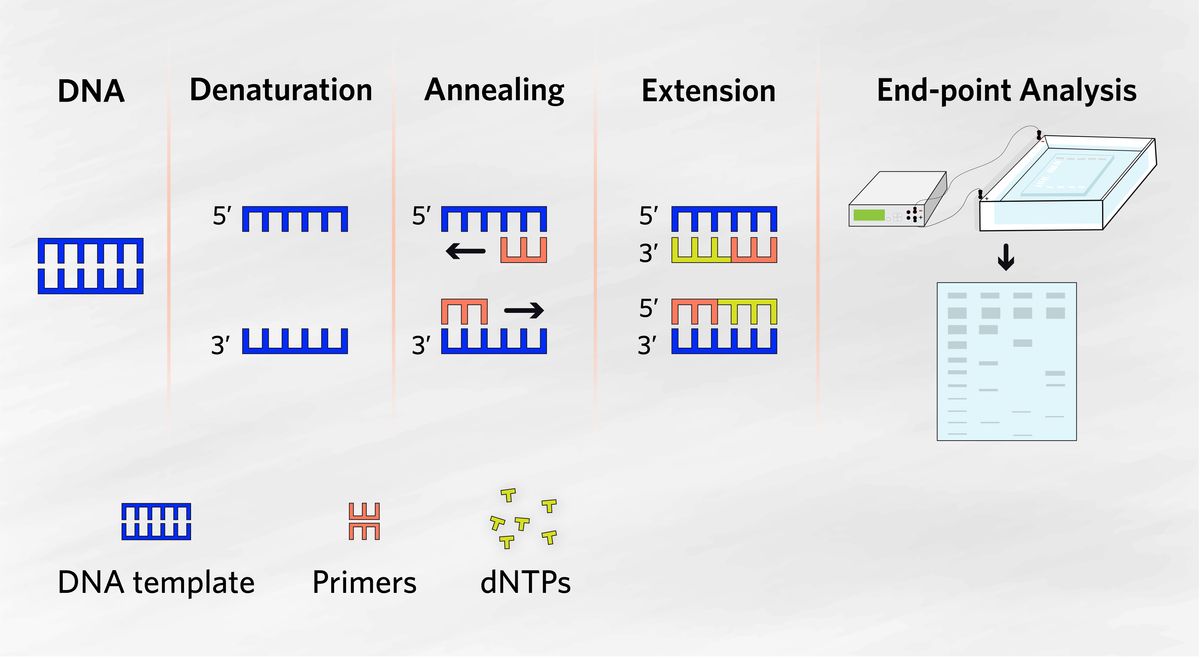
Polymerase chain reaction (PCR) is a fundamental molecular biology tool that scientists use to amplify and analyze genetic material, such as DNA and RNA. PCR involves the enzymatic amplification of nucleic acid templates, and can be divided into four major steps, listed below.1
- Denaturation: Double-stranded DNA separates into single strands.
- Annealing: Short DNA fragments bind complementary regions of the template to begin new strand synthesis.
- Extension: Nucleotides are added to the primers, which serve as extension points to continue new strand synthesis.
- Endpoint analysis: Scientists analyze PCR products typically using agarose gel electrophoresis.
Types of PCR
There are different types of PCR reactions based on their nature of amplification and detection methods. For example, end-point PCR is a qualitative method that identifies the presence or absence of the amplified target, but many quantitative methods also exist.2-4
Table 1: Types of PCR and how they compare to end-point PCR
Type of PCR Reaction | Description | Advantages |
Real-time PCR detects and quantifies DNA as the reaction progresses. | This quantifies the specific amount of DNA present in a sample. The procedure requires specialized thermal cyclers with fluorimeters. | |
Multiplex PCR uses multiple primers to amplify different targets in a single reaction. | This technique saves time and reagents by detecting several targets in a single reaction. | |
Low copy number PCR amplifies low amounts of DNA (typically <100-200pg). Increasing the number of amplification cycles to 34 instead of 28 helps increase sensitivity. Additional variations to the cycling conditions or PCR mix may also help with detection.5 | This method is particularly useful for detecting trace amounts of DNA, which makes it applicable in forensic investigations. | |
Complex genomic DNA (up to 20kb) and protein-coding DNA templates (30kb and more) can be amplified using long-range PCR.6 A combination of highly processive thermostable DNA polymerase and a high-fidelity polymerase with 3′-5′ exonuclease activity helps achieve long-range PCR.7 | This allows the amplification of long DNA molecules which cannot be amplified by regular PCR, especially genomic DNA. | |
PCR for cloning | PCR for cloning amplifies DNA fragments for ligation into a vector, either via a single base overhang or blunt end ligation. | This method allows cloning with low amounts of DNA and for high throughput applications. |
PCR for next-generation sequencing (NGS) | PCR amplification allows the creation of multiple NGS libraries. | This method facilitates sequencing multiple regions of interest at the same time. |
PCR Components and Tips for Optimization
Several components are involved in a PCR including template DNA, forward and reverse primers, DNA polymerase enzyme, and reaction buffer. Scientists must ensure that all reagents in the reaction are at optimal concentrations and ratios. Additionally, optimizing the thermal cycler conditions and using various additives can improve the PCR yield.
Template DNA
Template amount, source, and secondary structures or GC-rich regions may affect the reaction.7
- The amount of template depends on the DNA copy number. For a standard PCR reaction with 25-30 cycles, roughly 104 copies of the template DNA are sufficient to generate a detectable amount of PCR product.7
- The DNA required for a given reaction also depends on the source. Typically, 30-100ng of human genomic DNA is optimal for most PCR reactions. For more abundantly available genes such as housekeeping genes, 10ng is sufficient.
- Template complexity is important to consider before setting up a reaction. For templates with high guanine and cytosine (GC) content, scientists use a high melting temperature to separate secondary structures. Additives such as glycerol, dimethylsulfoxide (DMSO), and bovine serum albumin (BSA) can also help prevent secondary structures.
Forward and reverse primers
Primer design is extremely critical to a successful PCR reaction.7
- Optimal primer length is in the range of 15-30 nucleotides.
- GC content should be nearly 40-60 percent.
- Melting temperatures (Tm) should be between 52-58°C. The difference in Tm for both forward and reverse primers should not be more than 5°C.
- The 3′ end of each primer should preferably be a G or C base for strong hydrogen bonding and better annealing. However, this also increases the annealing temperature.
- Primer dimers are a common problem in PCRs. Primer dimers form by complementary primers self-binding instead of annealing to the template. Ensuring non-complementary 3′ primer ends helps prevent dimer formation.
- The optimal primer concentration range is 0.1-1μM. Higher concentrations could lead to primer dimers or non-specific template binding.
DNA polymerase
DNA polymerase is the foundational reagent for PCR. The Taq DNA polymerase enzyme, which researchers first derived from the thermophilic bacterium Thermus aquaticus, has played a significant role in standardizing PCR reactions.8 Due to its thermostable nature, Taq can withstand high denaturation temperatures and remains active in the incubator throughout the reaction cycles. Over the years, scientists have created several other recombinant DNA polymerases and explored multiple sources of polymerase enzymes to improve certain features for optimal PCR results.
Thermal stability: Archaea and bacteria that survive in extreme temperatures (above 80°C) are called hyperthermophiles. DNA polymerases derived from hyperthermophiles are very stable at high denaturation temperatures. For instance, the Pfu polymerase derived from the archaea Pyrococcus furiosus is more stable than conventional Taq polymerase.9
Several DNA polymerases with optimal activity at high temperatures can still function at lower temperatures, which can lead to non-specific amplification. Scientists prevent this by adding specific heat-labile antibodies that bind to polymerases and inhibit their activity at low temperatures.10 This is also referred to as hot-start PCR. Heat-sensitive chemical modifications to the enzyme's active site can also have a similar effect.
Fidelity: Scientists typically determine the fidelity of DNA polymerases by measuring their average error rate per base pair per duplication.11 Taq polymerase is known to be a low-fidelity enzyme and has an error rate between 2 × 10−4 to 2 × 10−5 errors/base/doubling. Most archaeal polymerases are high-fidelity enzymes because they possess 3'-5' exonuclease activity, which enhances the proofreading ability of the enzymes by allowing them to correct base mismatches. This is particularly useful for downstream PCR applications such as cloning, mutagenesis, or sequencing.
Extension rate: Most common polymerases perform nucleotide extension at 68-72°C with an extension time of one minute per ~1000 base pairs. Longer extension times may be effective for DNA templates longer than 3kb.7
Processivity: Polymerase processivity refers to the number of bases it can extend before detaching from the template. The structure of the DNA-binding domain largely determines this ability. Therefore, introducing mutations to polymerase DNA-binding domains can significantly enhance processivity.12,13
Additional PCR components
A typical PCR mix also contains a variety of other components that play an important role in ensuring an optimal reaction.7
- Magnesium salt (Mg2+) is an essential cofactor for thermostable DNA polymerases. The final concentration of Mg2+ in the PCR reaction mix usually ranges between 0.5-5.0mM.
- Deoxynucleotide 5′-triphosphates (dNTPs) are the building blocks of the reaction. Ideally, all four dNTPs should be present in equivalent concentrations, usually within the range of 20-200μM each.
- Additives are additional reagents that can modify and optimize PCR reactions.7
- DMSO can help lower the melting temperature (Tm) and optimize reactions involving GC-rich templates (typically >60 percent GC content). The recommended final concentration is 1-10 percent.
- Formamide weakens base pairing and increases primer annealing specificity, thus enhancing PCR amplification for templates with high GC content. Usually, scientists use a concentration range of 1.25-10 percent.
- BSA can alleviate the inhibitor effect of ferric salts and organic extracts present in certain water sources and biological samples, such as fecal matter. The optimal concentration in the reaction mix is around 400ng/μL.
- Non-ionic detergents such as NP-40, Tween 20, or Triton X-100 (0.1-1 percent) can stabilize DNA polymerases and prevent templates from forming secondary structures.
PCR cycling conditions
The steps of a PCR happen at different temperatures. Thermal cyclers capable of cycling through high and low temperatures drive these reactions.7
Table 2: Cycling conditions for a 3-step PCR
Step | Temperature | Time | Number of Cycles |
Initial denaturation | 94-98°C | 1 minute | 1 |
Denaturation | 94-98°C | 10-60 seconds | 25-30 |
Annealing | 52-58°C (5°C less than Tm) | 30 seconds | 25-30 |
Extension | 70-80°C | Variable* | 25-30 |
Final extension | 70-80°C | 5 minutes | 1 |
Hold | 4°C | Variable | 1 |
*Extension time depends on the amplicon length and the DNA polymerase.
Scientists modify the cycling conditions at each step to address challenges such as nonspecific amplification and low yield, improving reaction outcomes. One improvement is to extend the denaturation phase up to 5 minutes in cases where the starting DNA concentration is low, which helps prevent off-target annealing. Introducing the DNA polymerase when the reaction mix is heated to the denaturation temperature, as in hot-start PCR, is one way to extend this phase.
Assembling the reaction
There are several things to consider while putting together a PCR mixture, such as the stock solution components, optimal final reagent concentrations, and the order in which they should be pipetted into the reaction tube.1,7
Table 3: Typical PCR reagents and recommended final concentrations in a 50μL reaction mix
Reagent | Concentration in the Stock Solution | Final Concentration |
10X PCR buffer | 10X | 1X |
dNTPs | 10mM | 200μM |
MgCl2 | 25mM | 1.5mM |
Primers | 20μM (each) | 20pmol (each) |
DNA template | Variable | ~105 molecules |
Taq DNA polymerase | 5U/μl* | 2.5U |
*Taq starting concentration varies between manufacturers.
To streamline this process, researchers usually prepare a master mix containing reagents common to all samples (sterile water, 10X reaction buffer, dNTPs, and DNA polymerase). The sum of all reaction volumes, along with an additional 10 percent, is calculated and rounded up to the nearest whole reaction to determine the total volume for each reagent. For instance, if a researcher is setting up 10 reactions and needs to add 5μl of a 10X buffer to each reaction, the total volume of buffer required would be (10 reactions + 1) × 5μl = 55μl.
Troubleshooting Common PCR Challenges
Low or no amplification
PCR can suffer from low or no yield due to various factors such as incorrect reagent concentrations, inappropriate cycling conditions, or pipetting errors. To improve amplification, scientists add new reagents, prepare fresh working dilutions, set up appropriate controls, and tweak the PCR cycling conditions mentioned above. In some cases, the template might be old and may contain inhibitors that impede the reaction. This can be mitigated by incorporating additives such as BSA.14
Primer dimers and nonspecific PCR amplification
Primer dimers form by complementary primers self-binding. A simple way to reduce primer dimers is to modify the primer-to-template ratio. Suboptimal polymerase activity at low temperatures can cause nonspecific amplification. Hot-start PCR modifies the cycling conditions to help prevent both dimers and non-specific amplification. Nonspecific amplification from long genomic DNA can often lead to smear formation in PCR products, which can be resolved by performing PCR on a plasmid with the target sequence instead.
False positives
Given PCR’s high sensitivity, carryover contamination from previous products, cross-contamination in the laboratory environment, and repeat amplification of a target sequence can often lead to false positives. Several pre- and post-amplification sterilization techniques can prevent such contamination.15 For instance, mechanical barriers, such as PCR workstations that separate the sample preparation areas, and chemical barriers such as 10 percent bleach can effectively minimize contamination.
PCR products with sequence errors
Scientists need to consider sequence errors for applications downstream of PCR, such as cloning and NGS. DNA damage, base pair mismatches, or DNA polymerases with low processivity could all lead to erroneous sequences in the PCR product. Researchers can use single-molecule DNA sequencing to detect and address these errors.16
- Garibyan L, Avashia N. Polymerase chain reaction. J Invest Dermatol. 2013;133(3):1-4.
- Kubista M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27(2-3):95-125.
- Markoulatos P, et al. Multiplex polymerase chain reaction: a practical approach. J Clin Lab Anal. 2002;16(1):47-51.
- Keeney S. Long-PCR amplification of human genomic DNA. Methods Mol Biol. 2011;688:67-74.
- Gill P, et al. An investigation of the rigor of interpretation rules for STRs derived from less than 100 pg of DNA. Forensic Sci Int. 2000;112(1):17-40.
- Kee PS, et al. Long-range polymerase chain reaction. Methods Mol Biol. 2023;2967:181-192.
- Lorenz TC. Polymerase chain reaction: Basic protocol plus troubleshooting and optimization strategies. J Vis Exp. 2012;(63):e3998.
- Ishino S, Ishino Y. DNA polymerases as useful reagents for biotechnology - the history of developmental research in the field. Front Microbiol. 2014;5:465.
- Cline J, et al. PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24(18):3546-3551.
- Sharkey D, et al. Antibodies as thermolabile switches: High temperature triggering for the polymerase chain reaction. Biotechnology (N Y. 1994;12(5):506-509.
- Ricardo PC, et al. Fidelity of DNA polymerases in the detection of intraindividual variation of mitochondrial DNA. Mitochondrial DNA B Resour. 2019;5(1):108-112.
- Wu J, et al. DNA binding strength increases the processivity and activity of a Y-Family DNA polymerase. Sci Rep. 2017;7(1):4756.
- Wang Y, et al. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 2004;32(3):1197-1207.
- Chakrabarti R, Schutt CE. The enhancement of PCR amplification by low molecular weight amides. Nucleic Acids Res. 2001;29(11):2377-2381.
- Aslanzadeh J. Preventing PCR amplification carryover contamination in a clinical laboratory. Ann Clin Lab Sci. 2004;34(4):389-396.
- Potapov V, Ong JL. Examining sources of error in PCR by single-molecule sequencing. PLoS One. 2017;12(1):e0169774.

