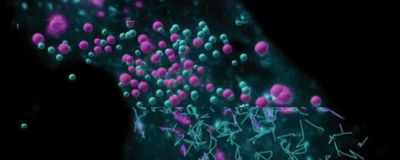
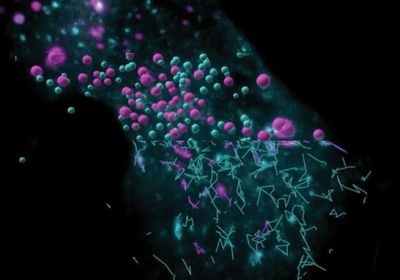
ABOVE: Endosomes within a single cell captured using lattice light-sheet microscopy, overlaid with their detected boundaries and trajectories. The Cellular Physiology Lab, Monash University
Endosomes, small lipid vesicles that sort and traffic biomolecules between other subcellular compartments and organelles, are critical to a cell’s internal transport network and overall function. However, the way that the endosomal system passes cargo from one endosome to the next in a timely manner was unknown. Harrison York, cell biologist at the Cellular Physiology Laboratory at Monash University and coauthor of a study published in Nature Communications, discussed how new technological advances in imaging such as lattice light-sheet microscopy (LLSM) and fluorescent lifetime imaging microscopy (FLIM) enable new approaches for visualizing and characterizing endosomal behavior.1
Why was it difficult to image endosomal movement?
The main limitation was time resolution because endosomes move incredibly quickly. For example, researchers using spinning-disk confocal microscopy could not image every endosome within a cell at a rate fast enough to visualize all of their movements. This allowed a rough understanding of endosomal maturation, but it was not possible to truly follow the process. They could not investigate how many endosomes mature per unit of time or look at higher-level things such as organization and regulation.
The other concern was photobleaching. Endosomes are quite small and every time they are illuminated there is a risk of bleaching the signals emitted by the labeling fluorophores. Furthermore, confocal microscopy illuminates—and thereby photobleaches—the entire sample during visualization, but can only capture a thin slice of the Z-plane at a time. This limits how long and, at the same time, fast one can record a phenomenon.
What is LLSM and what spurred your decision to use it?
LLSM is a technique developed by Nobel laureate Eric Betzig and his research team. Light-sheet microscopy uses a thin planar sheet of light rather than a single point to quickly scan a sample plane-by-plane. However, conventional or so-called “Gaussian light sheets” are too thick over cellular-length scales to resolve organelles and are better suited to studying larger structures such as organs inside developing animals. A thinner light sheet is required for subcellular imaging. Here, the Betzig group employed “beam-shaping” to create a structured light sheet roughly on the order of 400nm (instead of four to five micrometers for a Gaussian light sheet). LLSM enabled us to rapidly image every endosome within a cell at high resolution and over sufficiently-long periods so that we could accurately trace their trajectories, capture phenomena such as collisions between endosomes, and measure ensemble rates of conversion.
Why did you complement LLSM with FLIM?
FLIM let us infer whether two proteins were touching or not, which we used to determine which end of a protein was attached to a given vesicle. FLIM, in my opinion, is one of the more underutilized techniques in imaging. Whenever one excites a fluorophore, it enters an excited state, then drops to a ground state and emits a photon. This process typically takes a couple of nanoseconds, but is influenced by the local environment surrounding the fluorophore. Tracking these photon arrival times can provide information on the fluorophore environment. For example, when two spectrally overlapping fluorophores are within five to ten nanometers of each other, a Förster resonance energy transfer (FRET) interaction can take place which causes the second fluorophore to emit a photon. These FRET-derived photons can be detected because they shorten the overall average time it takes to receive a photon, and thus allows us to detect when proteins are directly bound to each other.
How can we use these imaging strategies and technologies to study other biological phenomena?
Biology is an incredibly rich space filled with dynamic processes, and the ability to image these processes has so much applicative potential. This study is only one example of how observing native intracellular processes can help uncover the mechanisms that govern the organization and robustness of life. Continual advances in microscopy have enabled scientists to image faster, longer, deeper, and with greater resolution. This is opening up the range of samples that can be observed, enabling us to ask questions regarding how these cellular dynamics operate in the local contexts of cells during development and disease.
This interview has been condensed and edited for clarity.
- York HM, et al. Deterministic early endosomal maturations emerge from a stochastic trigger-and-convert mechanism. Nat Commun. 2023;14:4652.