Bats have many adaptations, some of which are consistent across different species, that possibly contribute to their unusual resiliency to viral infections. However, bats are a heterogeneous group, with major lineages diverging more than 50 million years ago, so different evolutionary pressures may have given rise to important species-specific adaptations.
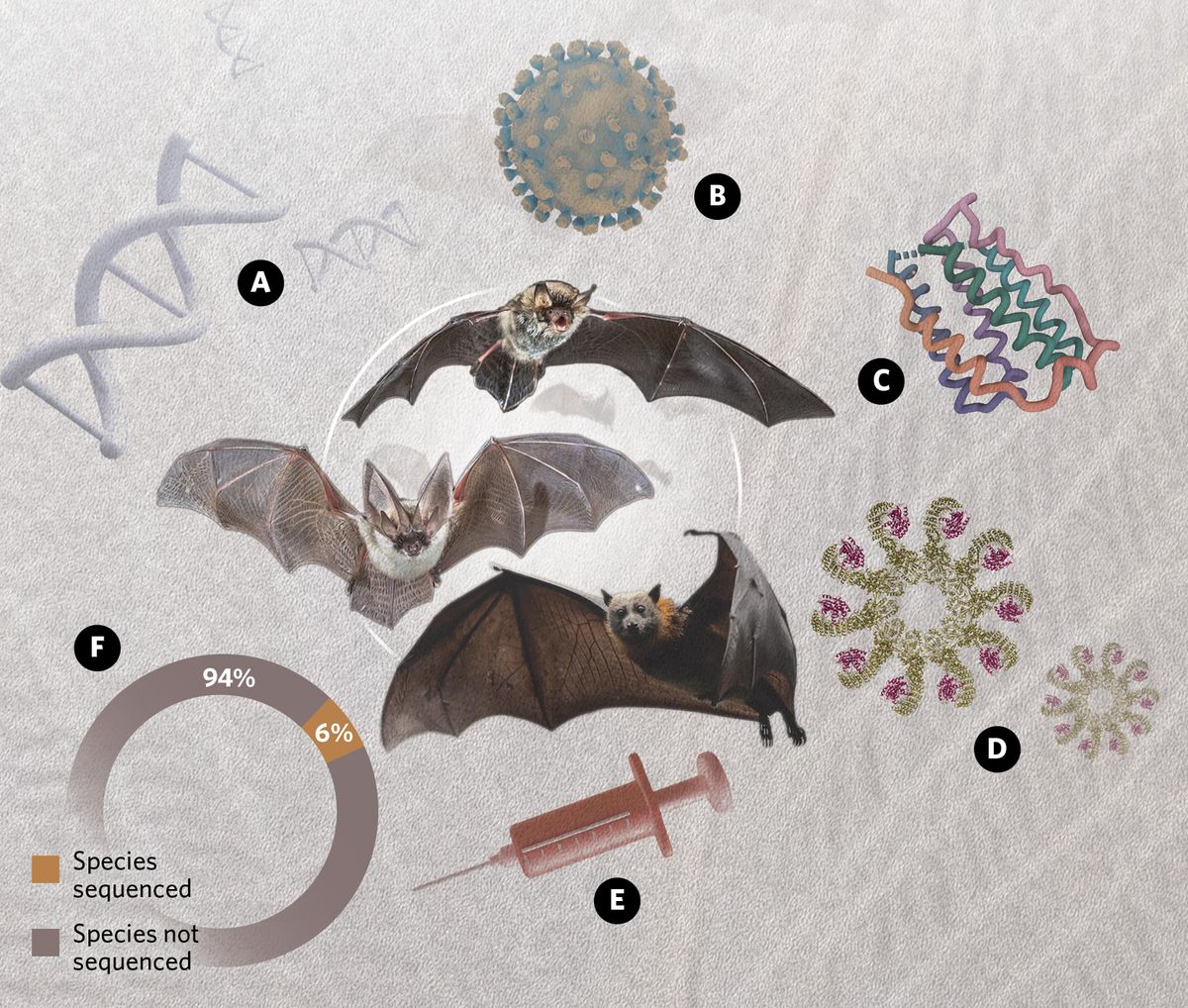
A) DNA Sensors
Several species of bats have lost the entire PYHIN gene family.1 Pyrin and hematopoietic interferon-inducible nuclear domain (PYHIN) proteins sense DNA, either from pathogens or the host’s own damaged DNA, and trigger an inflammatory response. Loss of this gene family may explain bats’ ability to limit harmful virus-induced inflammation.
B) Escape From Viral Antagonism
While some pathogen sensors have been lost, others have diversified. Protein kinase R (PKR) proteins are activated by double-stranded RNA, which signals the presence of a viral infection, and can then initiate an antiviral response. Many viruses, including some poxviruses, herpesvirus, and influenza viruses, have evolved ways to interfere with PKR function. Researchers think that PKR gene diversification may help bats escape this viral interference.2
C) Interferons
Interferons are key players in the antiviral response. Different bat species display differences in interferon genes and regulation of interferon expression. Some species show constitutive expression of interferon-α, while others have a notable expansion of the less studied interferon-ω genes.3,4
D) Inflammasomes
Bat ASC2 (apoptosis-associated speck-like protein containing a CARD 2) inhibits activation of the inflammasome. While inflammasome signaling is important for the antiviral response, overactivation has also been implicated in mortality for certain viruses, including influenza. Indeed, researchers showed that bat ASC2 increased the survival of influenza-infected mice.5
E) Possible Self-vaccination
A recent study on bat induced pluripotent stem cells revealed an unusually large number of endogenous viral sequences.6 While still unsubstantiated, the study’s authors suggested that future research should test whether these endogenous viral sequences serve to “defend against viruses and microbes and encode viral proteins in a self-vaccination scheme.”
F) Undiscovered Mechanisms
Bats are a diverse group with more than 1,400 species. According to approximate counts from the Bat1K gene sequencing initiative, very few species have fully sequenced genomes.7 Thus, many potential antiviral or anti-inflammatory mechanisms remain to be discovered.
- Ahn M et al. Unique Loss of the PYHIN Gene Family in Bats Amongst Mammals: Implications for Inflammasome Sensing. Sci Rep. 2016;6:21722.
- Jacquet S et al. Adaptive duplication and genetic diversification of protein kinase R contribute to the specificity of bat-virus interactions. Sci Adv. 2022;8(47):eadd7540.
- Zhou P et al. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc Natl Acad Sci U S A. 2016;113(10):2696-2701.
- Pavlovich SS et al. Egyptian Rousette IFN-ω Subtypes Elicit Distinct Antiviral Effects and Transcriptional Responses in Conspecific Cells. Front Immunol. 2020;11:435.
- Ahn M et al. Bat ASC2 suppresses inflammasomes and ameliorates inflammatory diseases. Cell. 2023;186(10):2144-2159.e22.
- Déjosez M et al. Bat pluripotent stem cells reveal unusual entanglement between host and viruses. Cell. 2023;186(5):957-974.e28.
- Sequencing status – Bat1K. https://bat1k.com/species-database/
Read the full story.
