Article reviewed by Govind Kaigala, PhD at the University of British Columbia.
Stay up to date on the latest science with Brush Up Summaries.
What Is In Situ Hybridization?
In situ hybridization (ISH) is a molecular technique that can localize and detect specific nucleotide sequences in cells, tissue sections, and even entire tissue.1 This technique relies on hybridizing the target nucleotide sequence with a complementary strand, known as a probe. Scientists label these probes with a radioactive, fluorescent, or chemical tag, which enables visualization of the probe binding with the target nucleotide.
The ISH Technique
Joseph G. Gall and Mary Lou Pardue, scientists at Yale University, performed the first successful ISH experiment in 1969 to identify highly amplified ribosomal DNA in Xenopus (frog) oocytes using a radioactive probe.2 Many years later, researchers advanced the ISH technique by developing fluorescent probes. ISH plays a vital role in cytogenetics, gene mapping, prenatal diagnosis, and gene expression.3
Tips and Tricks for Each ISH Step
The specific protocol researchers use for an ISH experiment depends on the type of sample, probe, and experimental conditions. “I would say there is almost no one fit solution for all,” said Govind Kaigala, a biomedical engineer at the University of British Columbia, who develops microanalytical techniques for quantitation in biology. Certain methodological tweaks, particularly during sample preparation, probe selection, probe labeling, and hybridization can improve ISH experimental outcomes.
Probe selection
In ISH experiments, researchers use different probe types such as complementary RNA (cRNA), complementary DNA (cDNA), and synthetic oligonucleotides.4 To select a suitable probe, scientists must consider multiple application-specific factors including sensitivity and specificity, ease at which the probe can enter the cell or tissue, probe–target hybrid stability, and method reproducibility.
Probe labeling
Before hybridization, scientists attach a label to the probe via nick translation or Taq DNA polymerase methods, which allows them to identify the specific location of the target nucleotide sequence in the cells or tissue sections. For probe labeling, researchers employ radioactive or nonradioactive techniques.4,5 Radiolabels such as 32P, 3H, and 35S exhibit high specific activity and allow scientists to detect transcripts present in low amounts.
Nonradioactive labels such as biotin, digoxigenin (DIG), and fluorescent molecules play an active role in ISH experiments. In comparison to the radiolabeling technique, nonradioactive detection methods are safer and more advantageous because they do not require radioactive materials handling. Furthermore, they do not involve hassles related to radioactive waste disposal and the probes labeled with these methods are more stable than 32P-labeled probes.
Sample fixation and preparation
Uniform, flat, and thin tissue sections or cell layers prevent uneven and variable background staining.6 Before performing ISH, scientists use a fixative to help preserve the morphology of fresh tissues or cells.7 For example, methanol/acetic acid solutions fix chromosomes in the metaphase stage of cell division, after the cell membrane is disassembled. Formaldehyde and Bouin's fixative are ideal for cryostat sections, while formalin is favorable for paraffin-embedded tissue sections. However, precipitating fixatives such as acetic acid and ethanol are less favorable for fixing fresh tissues because they make the cell matrix impermeable to labels and may modify the target nucleic acid, preventing or reducing hybridization.
After fixation, scientists section paraffin-embedded tissue samples using a microtome.8 Before conducting the ISH experiment, researchers dewax and rehydrate the sectioned samples.9 This allows uniform distribution of reagents on the tissue surface and ensures even staining.
Permeabilization
Permeabilization procedures are critical for removing proteins that surround the target DNA or RNA and allow probe diffusion through the cell.10 Fixatives used for sample preparation could cause protein crosslinking, which can mask the target nucleic acid. Scientists typically use proteinase treatment such as proteinase K or pronase enzyme, 0.2M hydrochloric acid, or detergents (e.g., Triton X-100) for permeabilization.11 Using the optimal concentration of these chemicals or enzymes is essential because a higher concentration may damage the tissue and disrupt cell integrity.
Prehybridization
Prehybridization steps help lower the background noise or nonspecific probe binding during ISH experiments.12 However, not all protocols require this step. This step is necessary when scientists use enzymes such as alkaline phosphatases and peroxidases to visualize labels, because the endogenous enzymes may increase the background noise. Peroxidase treatment can effectively reduce the background noise by quenching endogenous peroxidase activity and bleaching tissues. This treatment is particularly useful in fluorescence in situ hybridization (FISH) experiments using horseradish peroxidase (HRP)-labeled oligonucleotide probes.
Hybridization
Hybridization is a process in which the probe anneals with the complementary target sequence.3 The success of this step depends on hybridization solution composition, monovalent cation concentrations, pH, incubation temperature and time, organic solvents, and probe concentration. Researchers modify solution parameters to minimize nonspecific interactions in ISH and facilitate maximum on-target binding.
Hybridization efficiency is the rate limiting step of many ISH assays. Beyond solution parameters, scientists turn to new methodologies for optimal probe hybridization, such as microfluidic devices. In typical ISH experiments, the probe moves toward the target in the hybridization solution following a diffusion-based approach. This is a time-consuming step and the incubation time may reach up to 16 hours for low copy number targets and above 48 hours for entire genome hybridization. The time required for the probe to reach the target nucleic acid depends on the diffusion coefficient, which reduces with an increase in probe length.
Kaigala and his team addressed the problem associated with probe transportation in ISH experiments by bringing the probe towards the target, thereby reducing the overall assay time. “What we were championing is convective flows, so we are actively bringing the probe to the target,” said Kaigala. Efficient modification of a microfluidic environment can downscale reagent usage for ISH as well.13
During incubation, scientists monitor levels of hybridization solution to prevent it from drying via evaporation.14 This reduces the reaction volume and increases reagent concentrations. Post hybridization, scientists thoroughly wash their samples with buffer solution to remove nonspecific hybrids and unbound or loosely bound probes.
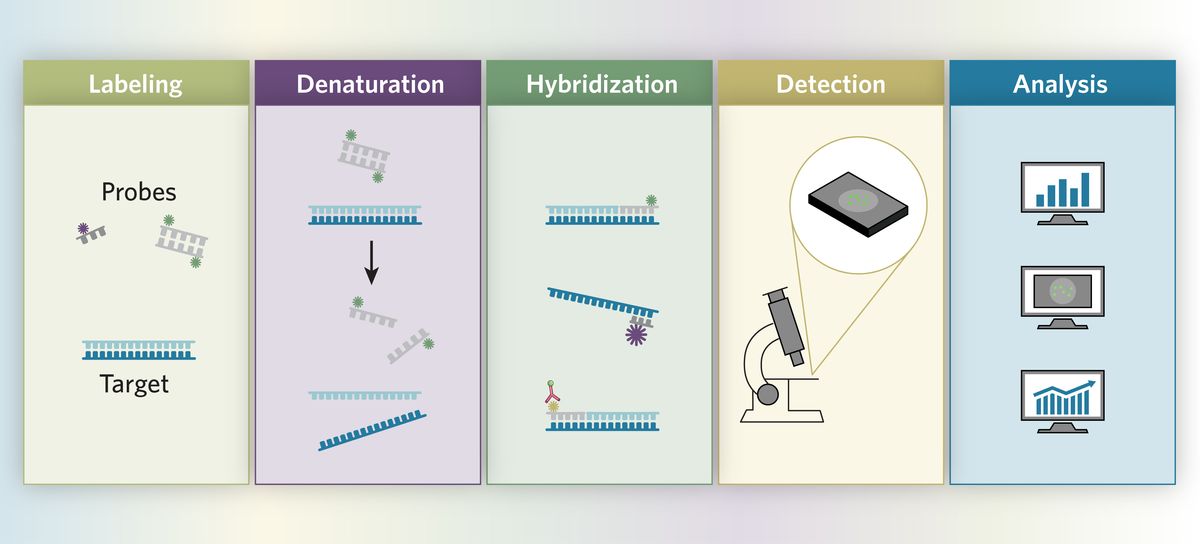
Detection
Many direct and indirect methods identify labeled probes. In the direct method, the reporter molecule (e.g., fluorescent marker or radioisotope) directly attaches to the probe. Researchers visualize the probe-target hybrids under a microscope, or with photographic emulsions or films immediately after hybridization washes. Indirect methods require additional steps for probe-target hybrid identification. Scientists add specific antibodies to identify biotin or DIG labeled probe-target hybrids.
ISH Methods
FISH
Fluorescence in situ hybridization (FISH) is a powerful cytogenetic technique that uses fluorescent probes to detect specific genes or chromosomes in cells or tissues.15 FISH allows scientists to visualize specific genes or portions of genes, and detect a variety of chromosomal abnormalities and genetic mutations. Researchers select from three different types of FISH probes, namely, locus-specific probes, centromeric repeat probes, and whole chromosome probes, based on their application.
Table 1: Common fluorescent dyes for labeling oligonucleotide probes16
Fluorochrome | Color | Excitation Peak (nm) | Emission Peak (nm) |
Alexa488 | Green | 488 | 496 |
CY3 | Red | 548 | 562 |
CY5 | Red | 651 | 670 |
AMCA | Blue | 356 | 434 |
Fluorescein | Green | 475 | 510 |
Rodamine | Red | 573 | 591 |
DAPI | Blue | 359 | 457 |
Kaigala’s research team optimizes the FISH technique with microfluidics. Using microfluidic devices, they actively deliver FISH probes to the cells or tissue samples. This strategy promotes constant probe replenishment and tracking hybridization kinetics in real time. “Typically, a FISH assay is an end-point assay,” said Kaigala. “We have the opportunity to check in real time how much of the hybridization has happened. We track the signal evolution.”
CISH
Chromogenic in situ hybridization (CISH) is similar to FISH but instead of a fluorescent dye, it uses chromogen.17 A chromogen is a colorless chemical compound that can be transformed into a colored form through a chemical reaction. In CISH, researchers use peroxidase- or alkaline phosphatase-labeled reporter antibodies, which interact with hybridized DNA probes and cause enzymatic reactions that allow scientists to detect the target by bright-field microscopy. CISH detects gene deletion, gene amplification, chromosomal number, and chromosomal translocations. The main advantage of the CISH technique is that signals do not diminish over time.
smFISH
Scientists use single-molecule fluorescence in situ hybridization (smFISH) to analyze gene expression in a cell by detecting and counting individual RNA molecules.18 This technique helps researchers investigate natural cell-to-cell variations in gene expression within a homogenous cell population.
MERFISH
Multiplexed error robust fluorescence in situ hybridization (MERFISH) can simultaneously image numerous RNA species in their native cell environment and spatially resolve single-cell transcriptomic measurements.19 This method helps map cellular diversity at a subcellular resolution and gene expression across whole tissues.
Challenges and Future Outlook
Variable and lengthy incubation times with unknown outcomes are additional ISH challenges. “We think there are a couple of ways forward. One is to see how to shrink the time, and at the same time, find ways to effectively multiplex,” said Kaigala.
To accomplish this, scientists need to fine-tune and customize existing ISH protocols. Over the years, specific research needs have governed the evolution of the ISH technique. For instance, researchers developed the smFISH technique to quantify the copy number of individual RNAs in a single cell.18 However, the limited number of RNA species that can be simultaneously imaged by smFISH motivated scientists to develop MERFISH.19
Challenges will continue to inspire new ISH methods. Currently, scientists need improved signal intensity for single RNA molecule methods. This is a limiting factor for a number of applications, particularly those related to imaging tissue samples that have a high degree of background noise and shorter RNAs.
- Veselinyová D, et al. Selected in situ hybridization methods: Principles and application. Molecules. 2021; 26(13):3874.
- Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. PNAS. 1969;64(2):600-604.
- Jensen E. Technical review: In situ hybridization. Anat Rec. 2014; 297(8), 1349-1353.
- Chu Y, et al. In situ hybridization: Introduction to techniques, applications and pitfalls in the performance and interpretation of assays. Semin Diag Pathol. 2019; 36(5).
- Chen CC, et al. Radioactive in situ hybridization for detecting diverse gene expression patterns in tissue. J Vis Exp. 2012;(62):3764.
- Libard S, et al. Characteristics of the tissue section that influence the staining outcome in immunohistochemistry. Histochem Cell Biol. 2019;151(1):91-96.
- Srinivasan M, et al. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161(6):1961-1971.
- Gracia AU, et al. Histological cut of a paraffin-embedded blastocyst: Optimized protocol for murine blastocysts. Methods X. 2019;7:100767.
- Redmayne N, Chavez SL. Optimizing tissue preservation for high-resolution confocal imaging of single-molecule RNA-FISH. Curr Protoc Mol Biol. 2019;129(1):e107.
- Rocha R, et al. Influence of the fixation/permeabilization step on peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) for the detection of bacteria. PLoS One. 2018; 5;13(12):e0208867
- Young AP, et al. A technical review and guide to RNA fluorescence in situ hybridization. Peer J. 2020;8:e8806.
- Lazar JG, Taub FE. A highly sensitive method for detecting peroxidase in in situ hybridization or immunohistochemical assays. In: Kessler, C. (eds) Nonradioactive Analysis of Biomolecules. Springer Lab Manuals. Springer, Berlin, Heidelberg.
- Huber D, et al. Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Micro Nano Eng 2018; 1, 15-24.
- May-Zhang AA, et al. Hybridization Chain Reaction for mRNA Localization in Single Cells from Mouse and Human Cryosections. Curr Protoc. 2022;2(5):e439.
- Chrzanowska NM, et al. Use of Fluorescence In Situ Hybridization (FISH) in diagnosis and tailored therapies in solid tumors. Molecules. 2020; 25(8).
- Bottari B, et al. Application of FISH technology for microbiological analysis: current state and prospects. Appl Microbiol Biotechnol. 2006; 73, 485–494.
- Paz N, et al. Combined fluorescent-chromogenic in situ hybridization for identification and laser microdissection of interphase chromosomes. PLoS One. 2013; 8(4):e60238.
- Chen KH, et al. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015; 348:6233
- Moffitt JR, Zhuang X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol. 2016;572:1-49.

