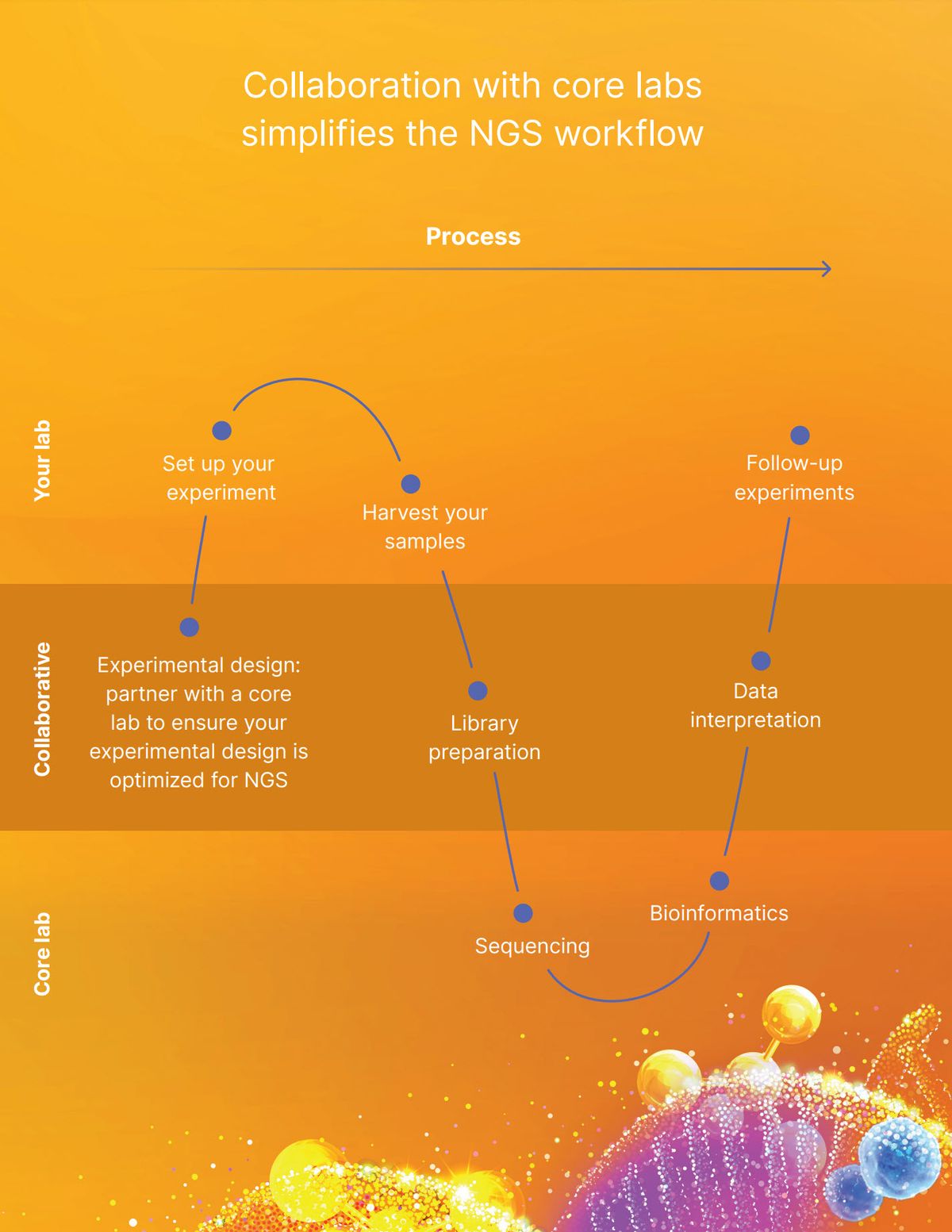
From bulk and single cell methods to spatial and multiomic methods, advancements in sequencing and -omics research are accelerating at an exciting pace. Next-generation sequencing (NGS) provides researchers from various scientific fields the ability to cast a wide net and explore complex pathways and biological processes. With NGS, scientists can take an untargeted approach, generating data from many genetic loci at once.
Such open-ended discovery allows researchers to see details that may be overlooked in targeted approaches that assay one gene at a time. Despite this benefit, the technical aspects of library preparation and data analysis can be daunting for researchers new to NGS. Previously, high sequencing costs, lack of expertise, and enormous data volumes were seen as barriers to entry.1 Over time, these barriers have diminished thanks to decreased costs and optimized workflows and bioinformatics pipelines.1-3 In addition, numerous academic NGS core facilities and commercial service providers have been implemented globally to support researchers on their NGS journeys. Such access to centralized equipment and expertise makes NGS more approachable to researchers new to the method.
Support for Scientists Diving into NGS
Instead of entering the NGS world alone, scientists can take their experiments to a core facility and receive expert guidance. “Our core motto is reducing barriers to genomic technologies. That means that whatever point you are in your knowledge of genomics, we want to be there to help you,” said Ann Tate, project manager for the Transcriptional Regulation and Gene Expression Facility (TREx) in the Biotechnology Resource Center (BRC) at Cornell University.
According to Tate, prior sequencing experience is not important when initiating an NGS project with a core facility. Some researchers may simply hand off their samples and wait for results, while others choose to take a hands-on approach, collaborating with core members from the onset of their project.
“We want to be a makerspace where people can come in, collaborate with us, get the training, have access to reagents, and actually get their projects off the ground and to the point where they have publishable data,” said Adrian McNairn, the lead biologist for the Genomics Innovation Hub in the BRC. From new graduate students lacking wet bench experience to tenured professors trying bioinformatics for the first time, researchers can work side-by-side with NGS core members who use their expertise to tailor technologies to specific research questions, train scientists in sequencing best practices, and make data analysis more accessible.
Core facilities and commercial service providers democratize NGS so that all scientists can access this powerful technology. Working with a core that has equipment such as sequencers, incubators, and fragment analyzers provides access in a way that is no longer cost prohibitive. “An example of a cool project that I did with someone who now has access…is iridescent butterfly wings,” said Tate. “We were sequencing butterfly wing cells. Five years ago, that would have been cost prohibitive to them.” Additionally, samples from lesser-studied species may require changes to existing reagents, workflows, or data analyses, which core members help customize thanks to their deep-seeded NGS knowledge. “We have seen everything from shark eyeballs, to pond scum, to broccoli and cauliflower,” Tate stated.
A hands-on, expert approach is also key when processing samples, building libraries, and analyzing data. As McNairn works with researchers, he evaluates the quality of their samples every step of the way. “Since we've worked with them through the whole process, we can go back and troubleshoot the areas that we know maybe went wrong,” said McNairn. In addition to support at the facility, core members facilitate connections between researchers and other resources on campus to help with projects that may be outside of the core’s expertise.
The holistic approach that follows a project from its inception to completion is a major benefit of working with a core lab. By receiving customized, expert support, even NGS novices can approach big research topics with confidence. “Now is the time to get started because the cost is right and because we have so much experience under our belt,” said Tate. “We can really facilitate any genomic experiment you want.”
NGS for All
Wet Lab Training for a Bioinformatics Expert
Some researchers may be experts in one area of NGS, but need support in another. That was true for Connor Kean, a graduate student in Andrew Grimson’s laboratory at Cornell University. Kean was experienced in bioinformatics when he first approached the NGS core at his institution, but was seeking help with sample preparation and library construction.
Kean studies how the immune system responds to its ever-changing environment, which requires exploring multiple -omes. In particular, Kean wanted to explore cells’ epigenomes in response to changing conditions by assessing genome-wide methylation patterns. Because he had no clear targets, an untargeted NGS approach that tested multiple loci at once best fit his research needs.
Kean was concerned that his lack of experience preparing samples for sequencing could lead to costly mistakes during his deep-sequencing experiment, but preparing his libraries side-by-side with core experts alleviated his fears. “Working with the core lab gave me the confidence to feel really good about not having to sink in a ton of time or money into something that was going to ultimately fail,” said Kean.
Through the hands-on work Kean did with support and training from the BRC, he generated sequencing data that he analyzed using his own bioinformatics expertise. Because he was involved in every step of the process, Kean was confident that his data was high quality. “That's why I wanted to work with the core lab to process the samples,” said Kean. “Knowing what I've done…makes the data analysis a lot easier on my own.”
Exploring Reproduction with NGS
In Paula Cohen’s laboratory at Cornell University, postdoctoral researcher Mercedes Carro and graduate student Amanda Touey study spermatogenesis. To understand the gene regulation that orchestrates this complex process, they used NGS to study the RNA from multiple cell types during spermatogenesis and narrow down targets for further research. “In my research, it makes sense to use untargeted approaches because we are studying Argonaute proteins, which are RNA binding proteins that regulate gene expression,” said Carro. “We want to be able to look at what's going on at the level of the genome.”
Because gene expression changes occur in multiple cell types throughout spermatogenesis, their project ended up being huge in scope, with mRNA and small non-coding RNA sequencing. “The small RNA sequencing was going to take a lot of customization and things that were well out of the reach of our capabilities,” said Touey.
Neither Carro nor Touey had previous NGS experience, so they met with BRC members to discuss the project’s background, the cell types and compartments they wanted to analyze, and the proposed protocol, after which they planned the sample collection strategy. “Talking to the core about [my] NGS goals and having them explain each step of the process made it more approachable to me,” said Carro. “Now I understand why it's important to process your samples in a certain way and why having certain controls [is important]. Not coming from a sequencing background, that helped build my confidence.”
The core experts also helped the researchers troubleshoot and avoid contamination while maintaining RNA integrity during sample collection. After sequencing, bioinformaticians built a custom bioinformatic pipeline for the small non-coding RNA analysis and reported the data in a way that was accessible to Carro and Touey. “Their expertise was able to find a really novel population of small RNAs that we would not have been able to find without them,” said Touey. “This really opened up a whole new area of research for us.”
While Carro and Touey started off being intimidated by NGS, the collaboration with the core facility taught them new skills that will support their future research endeavors. “I definitely feel like my confidence in bioinformatics has grown substantially,” said Touey. “I can't wait until I get to start my next project.”
By collaborating with core facilities, researchers new to NGS gain training and support while confidently tackling big questions and making significant discoveries. Download the new Introduction to Next-Generation Sequencing ebook from Illumina to learn more.
- Illumina. Benefits of NGS Targeted Resequencing. https://www.illumina.com/content/dam/illumina-marketing/documents/products/other/targeted-resequencing-guide-770-2016-012.pdf. Accessed June 20, 2023.
- National Human Genome Research Institute. DNA Sequencing Costs: Data. https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data. Accessed June 20, 2023.
- Dahlö M, et al. Tracking the NGS revolution: managing life science research on shared high-performance computing clusters. Gigascience. 2018;7(5):giy028.
