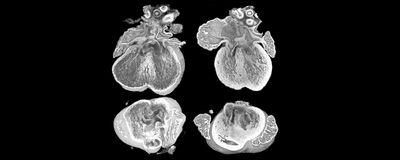
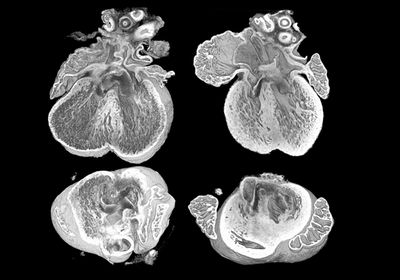
ABOVE: By analyzing images obtained with high-resolution episcopic microscopy, researchers found that an extra copy of the gene Dyrk1a contributed to heart abnormalities in a mouse model of Down syndrome. Eva Lana-Elola, The Francis Crick Institute
The making of the human heart is a complex process, and problems in heart development can result in congenital heart defects (CHD). The prevalence of CHD is close to one percent worldwide, but it rises to nearly 50 percent in babies with Down syndrome, a genetic disorder in which a person is born with a full or partial extra copy of chromosome 21.1,2
In a recent study, researchers sought to pinpoint the genes on chromosome 21 implicated in CHD. They found that three copies of the dual specificity tyrosine phosphorylation regulated kinase 1A (Dyrk1a) gene contributed to CHD in a mouse model of Down syndrome.3 These findings, published in Science Translational Medicine, uncover a gene potentially involved in heart defects in newborns with the chromosomal disorder.
“Until recently, we didn't know the identity of any of those genes for any of the conditions,” said Victor Tybulewicz, a molecular biologist at The Francis Crick Institute and author of the study. “Our work and the work of others is beginning to identify the causative genes for some of the aspects of the syndrome.”
In the late 1950s, researchers identified the trisomy of chromosome 21 as the genetic abnormality underlying Down syndrome.4 Forty more years passed before chromosome 21 was sequenced, revealing the genes that, when present in three copies rather than two, could contribute to health conditions associated with the genetic disorder.5
Over the years, scientists have attempted to map the genetic determinants of CHD on chromosome 21 using human samples and mouse models.6 Yet, pinpointing these dose-sensitive genes has proved a challenging task.
Nearly a decade ago, Tybulewicz and his team genetically engineered a mouse model, Dp1Tyb, with extra copies of 148 coding genes on mouse chromosome 16, which correspond to approximately 60 percent of the human chromosome 21 orthologues in the mouse genome. By engineering six other mouse strains with fractions of the Dp1Tyb duplication, the researchers identified a key region of the mouse chromosome 16 that links to CHD and found that at least two genes within that loci contribute to heart defects.7
“Having found that the mice did have this phenotype, we thought, ‘well this is an opportunity to go and find the genes that cause this,'" Tybulewicz said. To do so, he and his team first narrowed down the list of potential genes involved in CHD by comparing their previous findings with evidence from other researchers. They shortlisted five genes: Dyrk1a, SET domain-containing protein 4 (Setd4), carbonyl reductase 1 (Cbr1), microRNA 802 (Mir802), and potassium inwardly rectifying channel subfamily J member 6 (Kcnj6). Then the scientists reduced the number of copies of each of these five genes from three to two in Dp1Tyb embryos and used high-resolution episcopic microscopy (HREM) to obtain images of mouse hearts collected at embryonic day 14.5. “The 3D image is such a beautiful thing because you can rotate it on the computer in any direction you want and look at interior structures to see whether or not the heart has a defect,” Tybulewicz explained.
The occurrence of heart defects in the Down syndrome mouse model dropped to the level of control animals when the team decreased the Dyrk1a gene from three to two copies. To further investigate the effects of an extra copy of the Dyrk1a in the heart, the researchers compared the transcriptional profiles of wild type animals to Dp1Tyb mice with three or two copies of Dyrk1a. Compared to wild type animals, the hearts of Dp1Tyb mice showed a distinct transcriptional profile with downregulation of cell proliferation and oxidative phosphorylation genes, changes that resembled what the researchers found in human heart samples from fetuses with Down syndrome. In contrast, Dp1Tyb animals with two copies of Dyrk1a showed a gene expression profile similar to wild type animals, suggesting that an extra copy of Dyrk1a contributes to transcriptional changes in the hearts of the Down syndrome mouse model.
Since the transcriptomic analysis hinted at impaired cell proliferation and mitochondrial function in mice with an extra copy of Dyrk1a, the scientists next examined these processes more closely in heart cells. In cardiomyocytes and endocardial cells from Dp1Tyb mice, they observed disrupted cell proliferation with a high proportion of cells stuck in the G1 phase of the cell cycle. Reducing the dosage of Dyrk1a normalized the number of cells in different phases of the cycle, indicating that an extra copy of that gene is involved in impaired cell proliferation.
Cardiomyocytes from Dp1Tyb animals also had less elongated and branched mitochondria that showed reduced respiration rates compared to control cells. Decreasing Dyrk1a copy number reversed these morphological and functional changes, suggesting that three copies of the gene impair mitochondrial function in embryonic heart cells.
“They used this Down syndrome mouse model to do a pretty comprehensive study on heart defects,” said Kunhua Song, a heart development researcher at the University of South Florida who was not involved in the study.
Even though an extra copy of Dyrk1a associated with metabolic and proliferation changes as well as heart defects in the mice, it is still unclear whether these alterations have a cause-and-effect relationship. Tybulewicz hopes to determine this and to uncover the identity of other genes that may contribute to heart defects.
Song also noted that assessing how Dyrk1a interacts with these other genes in the context of CHD, as well as examining the contribution of this gene to heart defects in humans, would be important avenues to explore in the future. “Down syndrome affects a lot of people, and half of them are likely to have congenital heart defects,” he said. “It's definitely a big issue for this population.”
- Liu Y, et al. Global birth prevalence of congenital heart defects 1970-2017: Updated systematic review and meta-analysis of 260 studies. Int J Epidemiol. 2019;48(2):455-463.
- Santoro SL, Steffensen EH. Congenital heart disease in Down syndrome – A review of temporal changes. J Congenit Heart Dis. 2021;5:1-14.
- Lana-Elola E, et al. Increased dosage of DYRK1A leads to congenital heart defects in a mouse model of Down syndrome. Sci Transl Med. 2024;16(731):add6883.
- Antonarakis SE. Down syndrome and the complexity of genome dosage imbalance. Nat Rev Genet. 2017;18(3):147-163.
- Hattori M, et al. The DNA sequence of human chromosome 21. Nature. 2000;405(6784):311-319.
- Mollo N, et al. Genetics and molecular basis of congenital heart defects in Down syndrome: Role of extracellular matrix regulation. Int J Mol Sci. 2023;24(3):2918.
- Lana-Elola E, et al. Genetic dissection of Down syndrome-associated congenital heart defects using a new mouse mapping panel. Elife. 2016;5:e11614.