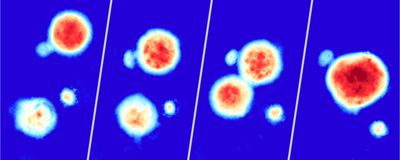
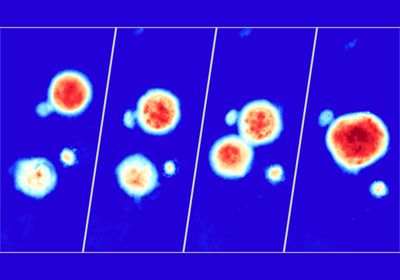
ABOVE: Alice Soragni and her team used their high-speed live cell interferometry-based platform to acquire images like this one, which displays bioprinted cancer cell line clusters over the course of 72 hours. Peyton Tebon and Bowen Wang
The success of nonclinical drug testing relies heavily on using models that accurately recapitulate complex biological processes. With the passing of the Food and Drug Administration (FDA) Modernization Act 2.0 in 2022, researchers are no longer required to employ animal models to screen drug candidates.1 Instead, the FDA is encouraging scientists to use other models, such as three-dimensional organoids and artificial intelligence-based methods, which are more predictive of the human response.2 As a result, researchers will need to improve existing methods and develop new protocols to optimize the efficiency of nonclinical testing. In a proof-of-concept Nature Communications paper, scientists developed an automated high throughput assay to monitor tumor organoid growth label-free.3
“3D screening in a high throughput manner is still very challenging,” said Alice Soragni, a cancer biologist at the University of California, Los Angeles (UCLA) and co-corresponding author of this study along with Michael Teitell at UCLA. Normally, researchers manually seed organoids within a 3D extracellular matrix, such as Matrigel, which is either spread across the surface of a microplate’s well or placed as a drop in the center of the well. However, this methodology prevents scientists from implementing automated liquid handlers, which would disturb the organoids grown in the center of each well. To overcome this hurdle, Soragni, Teitell, and their interdisciplinary team developed a strategy to bioprint cancer cell line clusters suspended in culture medium and Matrigel into a microplate’s wells. The bioprinter extruded the mixture in the shape of a hollow square, leaving a clearing in the center of each well. Because they could now employ automated liquid handlers, the researchers could increase the pipeline’s throughput.
The researchers also wanted to develop a method for observing how individual tumor organoids respond to therapy in real time. Scientists commonly use endpoint assays to assess a drug’s effect on an organoid population, but these tests fail to evaluate intra-sample heterogeneity. Soragni and her colleagues instead employed high-speed live cell interferometry, which is a label-free optical technique that uses phase shifts generated when light passes through an object to calculate its mass. Using this imaging method and machine learning-based quantitation, the researchers evaluated how individual tumor organoids responded to treatment with anticancer drugs by monitoring their biomass. Cell populations arising from the same line are often assumed to be homogenous, but after treatment the researchers observed organoids increasing, decreasing, or maintaining their size. These findings highlighted the pipeline’s potential for detecting heterogeneous drug responses.
“It is very nice to have an interdisciplinary group working together to figure out what we can do better,” said Lanlan Zhou, a cancer biologist and the director of the Organoid Core Facility located within the Legorreta Cancer Center at Brown University, who was not involved in the study. Zhou was excited to see what the group would do next and hoped they would test their platform’s ability to assess more complex organoid models that better represent the tumor microenvironment in vivo. “You do have to think about the tumor microenvironment because the tumor is not only made of tumor cells. It is made of many cells including some normal cells. For example, they have immune cells, they have fibroblasts, they have blood vessels.”
Soragni, Teitell, and their team are currently investigating how patient-derived organoids will react to bioprinting in hopes that they can use this platform to predict patient drug responses. This will be of particular importance for rare cancers, such as sarcomas. “For the majority of rare cancers, we have very little knowledge. We do not know what are the drivers of disease, we do not know how heterogeneous they are, we do not know how they respond to therapy,” Soragni said. “Looking with this type of platform at rare cancer is going to give us a lot of information. It is going to teach us a lot of biology and it is going to also help us potentially come up with therapies we could be looking at clinically.”
- Adashi EY, et al. The FDA Modernization Act 2.0: Drug testing in animals is rendered optional. Am J Med. 2023;136(9):853-854.
- Zushin PJH, et al. FDA Modernization Act 2.0: Transitioning beyond animal models with human cells, organoids, and AI/ML-based approaches. J Clin Invest. 133(21):e175824.
- Tebon PJ, et al. Drug screening at single-organoid resolution via bioprinting and interferometry. Nat Commun. 2023;14(1):3168.