ABOVE: © SHUTTERSTOCK.COM,
NOBEASTSOFIERCE
A few minutes from the University of Pennsylvania campus in Philadelphia sits a small peach orchard that’s home to some unusual experiments. Contrary to first appearances, the subjects of the experiments are not the peach trees themselves, each of which is protected by a two-meter-cubed tent of fine mesh material. Instead, researchers are interested in the hundreds of tiny fruit flies living on the trees and the even tinier bacteria living inside the insects’ guts.
The setup was designed with a deceptively simple question in mind: Do the microbes in an animal’s digestive tract help shape their host’s evolution? Evolutionary biologist Seth Rudman says that it would make sense if they did. “Microbiomes [can have] a huge effect on host fitness, and hence could have a huge effect on adaptive trajectories of populations,” says Rudman, who helped construct part of the site in 2017 while a...
Rudman set out to do just that, in an outdoor environment subject to wind and rain and other disturbances from the real world. In what he describes as a “first pass” at this kind of research, the team released a thousand Drosophila melanogaster into each peach tree enclosure—cleared of ripening peaches so that the flies would have to eat whatever the researchers provided them. Then, the team added plates of food: control fly populations received standard food, while others got food laced with one of two bacteria commonly found in the insects’ guts, Acetobacter and Lactobacillus. “By doing that, we were sort of able to move their microbiomes towards an Acetobacter-dominated [composition] or a Lactobacillus-dominated [one].” The fruit flies were left to reproduce for five generations before Rudman went back to the orchard to collect some of the insects for genetic sequencing.

In 2019, the team published its findings showing clearly that, as Rudman had hypothesized, the populations of Drosophila had diverged from one another, with the several Lactobacillus-fed populations showing different allele frequencies from the populations that had been fed Acetobacter. (See illustration.) “When you have population changes in parallel [like that], that’s amongst the strongest evidence for adaptation,” explains Rudman, who recently started his own lab at Washington State University. Although he and his colleagues didn’t dig into what the genetic variation meant for the flies’ phenotypes, they did show that it mirrored the variation observed in wild Drosophila populations across geographic latitudes in North America—for instance, alleles associated with a Lactobacillus diet in the experiment are more frequent in populations living at higher latitudes, where these bacteria are more abundant. That really “keyed us in that [the genetic divergence] was meaningful,” Rudman says.
The study is “probably the cleanest demonstration of a host adaptive response to the microbiome” so far, says Andrew Moeller, an evolutionary biologist at Cornell University who is working on similar questions in mammals and was not involved in the Penn team’s research. “It strongly suggests that the latitudinal gradient in Drosophila allele frequencies is driven at least in part by the different microbes they’re exposed to with latitude.”
The findings fit with a growing literature about the gut microbiome’s role, largely overlooked until now, in shaping species’ genetic past and future. Although few organisms are as practical to study as fast-growing Drosophila, research on other species in the past decade or so has produced multiple reports of common phylogenetic patterns in animals and their microbial residents—a phenomenon known as phylosymbiosis—and scientists have documented various correlations between microbiome composition and host phenotypes. In many cases, these studies have revealed tantalizing clues of microbiome-driven evolution in animals, including humans, and in some cases, pointed to specific adaptations that likely owe their origin, at least in part, to bacteria in the digestive tract. As Rudman puts it, “there’s enough data now, both from phylosymbiosis, patterns of intraspecific variation in microbiome composition, and experiments like the one we did in 2019, that suggest it is important and it should be looked at more deeply.”
Dependent relationships
Ruth Ley did her PhD in evolutionary biology, so when she started getting involved in microbiome research as a postdoc in Jeffrey Gordon’s lab at Washington University School of Medicine in the early 2000s, she couldn’t help but wonder about gut microbes’ evolutionary histories. “We spent a lot of time looking at the [phylogenetic] trees” of microbes in the mammalian gut, she says. “That’s when I really began thinking about how they could have coevolved” with their hosts.
Microbiologists were already studying coevolutionary relationships between microbes and hosts back then, but typically in pathogenic settings or other relatively extreme biological associations—for example, that of aphids and the symbiotic bacterium Buchnera aphidicola, which synthesizes amino acids critical for aphid metabolism. These kinds of paradigms didn’t fit particularly well with what was known of mammals and many other animals, which incubate vast numbers of typically innocuous resident bacterial species, none of which is strictly essential to host survival.
It’s precisely what you would expect if there’s been this history of adaptation of the host to its microbiome.
—Andrew Moeller, Cornell University
Interested in whether coevolution might be occurring in these kinds of biological partnerships, Ley and colleagues decided to take a look at the whole mammalian clade. They developed phylogenetic trees for gut bacteria sampled from 60 mammalian species including humans, and overlaid these trees onto the phylogenetic tree for mammals. The findings, published in 2008, showed corresponding patterns between the trees, with the nodes and branches in fecal bacterial lineages tending to match those of the mammalian tree, suggesting that many bacterial lineages have diversified alongside their mammalian hosts over millions of years.
More-recent studies have borne out similar relationships, says Moeller. Phylosymbiosis “seems to be kind of true across many mammalian clades, where the composition, and also . . . the evolutionary history of the individual lineages in the microbiome, reflects the host phylogeny.” Using fecal samples from chimps, gorillas, and bonobos combined with existing data from humans, for example, he and his colleagues showed a few years ago that bacterial phylogenies closely aligned with that of primates, suggesting that at least some microbial lineages have been associated with humans “since our common ancestors with African apes,” Moeller says. “It means many of our microbes are evolutionarily ancient.” Ley, now at the Max Planck Institute for Developmental Biology in Tübingen, and others have since studied various vertebrate taxa and reported that signals of cophylogeny seem to be particularly strong in mammalian clades.
Of course, while a long-term relationship between a host species and its microbiome suggests that something is maintaining the association, it doesn’t mean that the former has adapted in response to the latter. There are other, non-exclusive processes that could lead to similar patterns, Moeller says: microbes in the gut might be adapting to their hosts; hosts and bacteria may share evolutionary trajectories because they’ve been evolving in the same environments; or patterns may arise simply from the accumulation of neutral mutations over time.
Evolution experiments aren’t nearly as feasible for mammals as they are for Drosophila, so to disentangle these potential explanations, scientists have paired phylogenetic research with microbiome-swapping experiments—the prediction being that if an animal species has undergone adaptation to a particular microbiome, then a host should have a higher biological fitness when paired with its own microbiome than when supplied with that of another species. Several studies are consistent with this idea. As early as 2012, for example, a team at Harvard Medical School reported that mice reared with a human- or rat-specific microbiome failed to develop the full complement of immune cells and were more vulnerable to infection than mice with normal murine gut bacteria.

More recently, Moeller and colleagues swapped microbiomes among house mice and three closely related mouse species and found that the animals fared better when paired with their normal microbiota, growing faster and showing lower levels of inflammation-associated proteins. The magnitude of the effects corresponded to the evolutionary distance between donor and recipient. In other words, the microbiomes of “distantly related donors had more of an effect on the house mice, and [those from] more closely related mice had less of an effect,” Moeller explains. “It’s precisely what you would expect if there’s been this history of adaptation of the host to its microbiome.”
While most experiments have focused on rodents, several researchers have argued that humans, too, have likely adapted to their microbiomes over evolutionary history, and that recent rapid changes in the composition of human gut microbiota could be responsible for some of the health problems increasingly observed in Western industrialized societies. Stanford University microbiologist Justin Sonnenburg, for example, proposes that the rising incidence of inflammatory diseases such as metabolic syndrome and obesity is partly a result of a “mismatch” between people’s current microbiomes and those that they have evolved with over the last few million years. That’s a hypothesis for now, he notes. Indeed, some researchers have recently advised more caution in interpreting the microbiota’s role in human disease, given that most studies on the topic are based on observational data and animal research. Several medical-focused microbiome research groups are now working to close this gap.
Animals harbor vast numbers of microbes in their guts, but the effect of this community on the evolutionary trajectories of the hosts is unclear. Researchers are now using a combination of experiments and observational studies to look for signs of microbiome-driven host adaptation. |
Rapid evolution in an outdoor experiment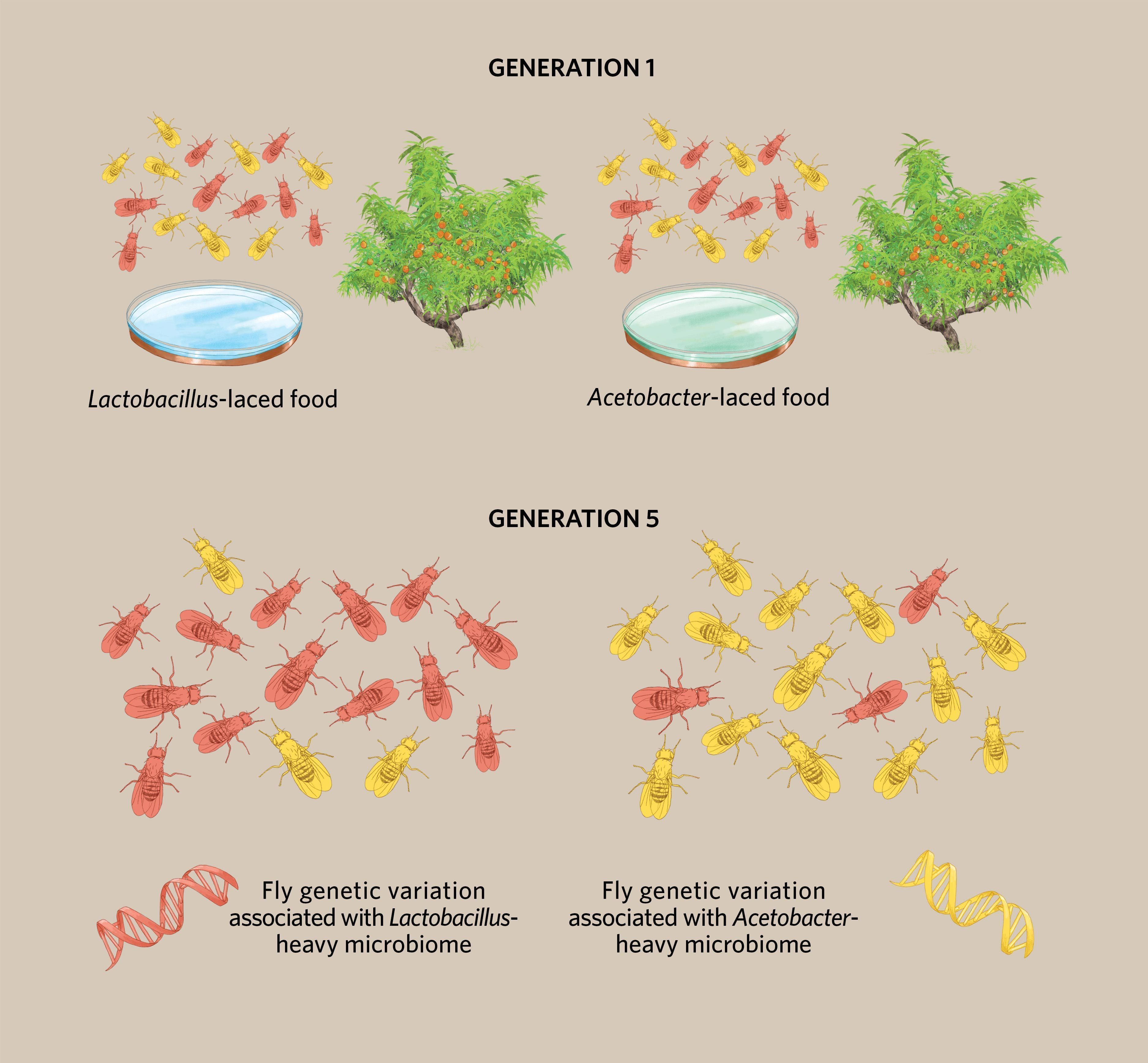 © Michelle Kondrich Researchers at the University of Pennsylvania split a large outbred population of fruit flies (Drosophila melanogaster) into 14 groups of 20 individuals and put each group on a peach tree enclosed with a mesh cage. Each population was given Lactobacillus-laced food (left), Acetobacter-laced food (right), or food with no bacterial addition (not shown). After five generations, the team collected the flies for genomic sequencing and found that the groups differed depending on the food they’d been given. (Graphic illustrates one possible scenario for variation at a single locus.) Some of the observed changes in allele frequencies matched variation seen in wild fruit fly populations whose diets have different concentrations of Lactobacillus and Acetobacter species, suggesting the microbes might help drive adaptive evolution in the real world, too. |
Microbiome-swapping among closely related species © Michelle Kondrich Evolutionary biologists predict that if an animal population has adapted to its gut microbiomes, then individuals in that population should suffer reduced fitness when given unfamiliar microbiomes. Researchers recently reported evidence consistent with this prediction using related species of mice. Mus musculus domesticus had higher fitness (as measured by growth and a low rate of inflammation) when implanted with microbiomes transplanted from other M. musculus than with microbiomes from three closely related species, and an individual mouse’s fitness correlated with the degree of relatedness to its donor species. |
A potential microbiome-influenced adaptation in humans © Michelle Kondrich The ability of adults to digest lactose is a uniquely human trait, and some researchers theorize that its evolution may have been influenced by the gut microbiome. Like other mammals, ancestral humans would only have produced the lactase enzyme in infancy. However, unlike other mammals, these humans at some point started consuming milk in adulthood. Initially, the theory goes, they would have had lactose-digesting bacteria in their guts that broke down the sugar and used most of the energy released as a result, leaving only a little for the host. In the event that food other than milk became scarce, humans would have been under intense selection pressure to get more energy from this food source, meaning that any mutation that gave adults the ability to digest their own lactose would likely have been favored. Lactose-digesting bacteria might subsequently become less abundant in the guts of these people, as they now have to compete with the host for food—a pattern that researchers say matches with some observations of current human populations. See full infographic: WEB |
Stepping stones
Another way to learn about how resident bacteria could shape species’ evolutionary trajectories is by thinking about the specific sorts of adaptive host genetic changes that might be driven by host-microbiome interactions. For example, if the hypothesis Sonnenburg describes about a change in human guts leading to more-common inflammation turns out to be correct, human genomes could experience selection to minimize the harmful effects of that change, he speculates. “You can imagine over time that we would select against certain alleles in our genome that are pro-inflammatory. Over time, we would settle into a new human genome-microbiome equilibrium.”
Researchers are also interested in cases where the microbiome might have helped their hosts gain new functions or exploit new environments or niches—digesting a particular novel food source, say, or defending themselves better against a particular pathogen. Indeed, it’s well-known that microbes, simply by performing certain chemical functions in the gut, can expand an organism’s dietary niche. A famous example is the desert woodrat, which is able to consume highly poisonous plants thanks to certain microbes living in its gut. Koalas’ specialized diet of eucalyptus, too, is likely aided by bacteria that detoxify the plant in the animals’ gastrointestinal tract.
Just how this kind of microbiome-enabled niche expansion influences genetic evolution of the host is a trickier question to address, says Moeller. In theory, having a microbiome that buffers the host against a new habitat or environmental change “can actually slow down the adaptive response,” he says. “Another way to say it is that if there’s a new environmental challenge and your microbiome can change very rapidly to eliminate any negative effects on fitness of the new environmental challenge, that removes all selective pressure on the host population to adapt [genetically].”
While most experiments have focused on rodents, several researchers have argued that humans, too, have likely adapted to their microbiomes over evolutionary history.
Over the longer term, though, a microbiome-provided ability to exploit a new resource or survive in a new environment may have rather different consequences for host evolution. It could permit dietary diversification, for example, and thus promote coexistence among species that would otherwise compete with one another for resources. Researchers in the US and in Madagascar have invoked this hypothesis to help explain the coexistence of four closely related lemur species: different gut microbiota assemblages help the species digest particular foods and promote distinct behaviors, helping maintain species diversity on the island, the team suggested in a recent study. In other kinds of scenarios, Ley says, the microbiome may act as “a stepping stone”—initially providing a function to the host before that function is taken over by the host genome, perhaps when it becomes beneficial for the animal to do it itself.
Ley has been studying possible cases of this latter phenomenon in humans, and tells The Scientist that the most compelling example involves the persistence of the lactose-digesting enzyme lactase into adulthood. “Lactose in the diet as an adult is a very human, weird aberration,” says Ley. Most mammals only need to digest milk when they’re infants, after which expression of the LCT gene, which codes for lactase, is switched off. But a mutation in LCT in several human populations, particularly in northern Europe, causes the gene to be expressed into adulthood. Ley and others have proposed that the human microbiome played a significant role in this change.
According to the researchers’ hypothesis, early human adults lacking the lactase enzyme likely would have been able to consume lactose and even get some energy thanks to gut microbes that digested the compound. “You’ve got microbes that can just ferment it like any other sugar,” Ley says. Somewhere in our history, the theory goes, some human populations suddenly became much more dependent on milk as a food source. At this point, Ley says, the host would gain an advantage by digesting its own lactose—it would obtain more energy. “Certain people who were just by chance lactase persistent, they out-survived the others, and you have the host genotype taking over from the microbes.”
Several observations of current European populations align with this hypothesis. For example, people with LCT mutations that cause persistent expression of lactase typically have lower abundance of certain lactose-digesting bacteria in their guts compared with people who don’t make lactase as adults, supporting the idea that there’s competition between host and microbes for this resource. Ley and her colleagues are now gathering more data on the relationships between host genotype, milk consumption, and microbiome composition from human populations around the world.
The team has pursued a similar line of thinking for starch digestion. Studies of modern humans suggest that genes coding for enzymes that break down starch underwent duplication sometime after humans split from Neanderthals, Ley notes, and current human populations show the same kinds of correlations between genotype and microbiome composition that they do for lactose digestion. She suspects there may be more examples worth exploring, too; a few months ago, she and a colleague reviewed various traits for which humans and microbes function via the same mechanisms—by metabolizing the same dietary molecules, for example, or by inducing the same physiological pathways—and proposed that they could all theoretically represent opportunities for microbes to influence host evolution.
These kinds of observational studies can’t demonstrate a causal relationship between variation in host genomes and the microbes associated with that variation, Ley says. But just describing the correlations is a start, she adds. “We think of it as a roadmap to interrogate how [humans and their resident microbes] might have interacted. It’s pretty wide open—I think people are just starting to think this way.”
A sharing community
There’s still a number of unanswered questions about how hosts and microbiomes interact—and about how they can best be studied. For example, while biologists often talk about microbiomes in terms of species composition, some argue that it would make more sense to focus on the functions of bugs in the digestive tract rather than the taxa those bugs belong to. Gut bacteria frequently exchange genetic material, such that one bacterial species within one microbiome might behave very differently from that same species in another microbiome. “The microbiome isn’t a fixed thing,” Rudman says. “It’s a set of genomes that are interacting and changing in frequencies. The complexity of the question is what is truly fascinating—and sometimes daunting.”
Another important consideration is how particular microbiomes are maintained across generations in a population, Moeller notes. Perhaps the most obvious way is vertical transmission—that is, inheritance of microbes from parent to offspring. Maria Gloria Dominguez-Bello’s lab at Rutgers and others have described how vaginal birth in humans helps shape the microbiome of infants, for example. And experiments by Moeller’s group have shown that, in lab mice, vertical transmission is the main mode of transfer for almost all of the gut microbiota.
There are other ways that microbiomes could be maintained across generations, even without being passed 100 percent reliably from parent to offspring. One way is if heritable behaviors or gut morphologies make a host more likely to acquire or retain certain microbes from the environment, for example. Another idea is that offspring could reliably inherit or pick up just a few key microbes from their surroundings, and then have those microbes subsequently shape the rest of the community.
Joy Bergelson, an ecologist and evolutionary biologist at New York University, has pursued this idea with the microbiome of the model plant species Arabidopsis thaliana. Using wild Arabidospsis populations, her team showed that around 10 percent of microbial taxa are highly conserved across generations, and that these particular microbes seem to occupy important positions, or hubs, in the network of microbial interactions taking place in the plant microbiome. The team doesn’t know yet how those “hub” microbes are maintained—it’s likely that the plant picks them up selectively from the environment, Bergelson says, though some researchers have suggested that microbes might also be inherited directly through seeds. In any case, “it gives you this picture of the plant actually controlling the microbiome by controlling the hubs,” says Bergelson, whose team posted some of the results as a preprint a few years ago.
It’s not clear whether microbiome inheritance might work this way in animals, but it’s “a fascinating concept,” Rudman says. Offspring would potentially only need to inherit a small community of microbes from their parents, and then “the microbe-microbe interactions that happen within the gut could essentially lead to a deterministic outcome that produces a very similar microbiome across generations.”

Understanding these kinds of dynamics is challenging in the limited environment of a typical lab, so researchers are increasingly taking their experiments outside. Moeller’s group, for example, has started using a large outdoor enclosure in which radio-tagged mice can scamper about and interact as they please. “We hope it’s a way to tease out the various transmission modes of the microbiome—through social networks, through environmental transmission, or through vertical inheritance,” Moeller explains. The first dataset from that project has just recently come in for analysis, he says, adding that the team plans to use the enclosure setup to address longer-term questions about microbiome-related adaptation, too.
Rudman, meanwhile, has been carrying out more research in Drosophila and also plans to work with stickleback fish, a species commonly used in adaptive evolution studies, he says. Designing experiments that capture as much of a species’ ecology as possible will be particularly important, he adds, not only for understanding how microbiomes influence host genomes, but for determining the extent to which this influence matters, among all the other forces at play, in driving host evolution in the real world. “I think the jury’s out on that,” he says. “The data will hopefully guide us—and that’s the way science should go.”
Interested in reading more?







