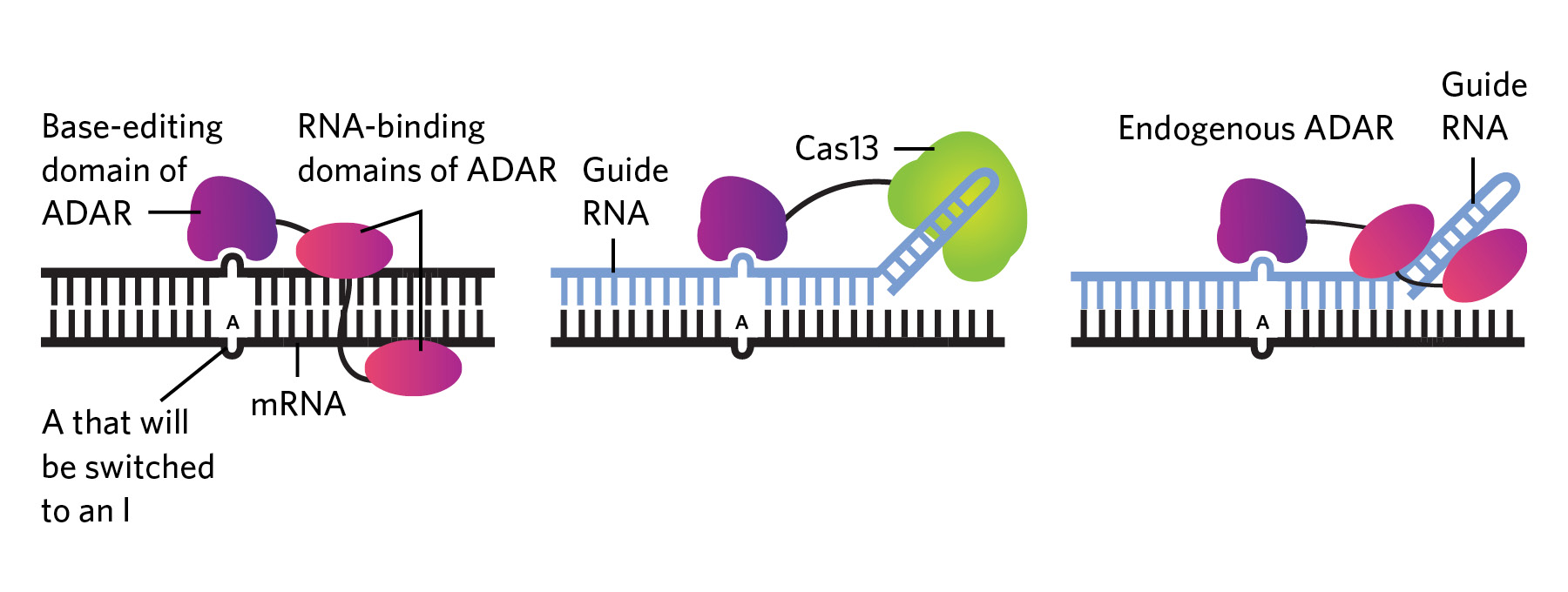
Editing RNA isn’t all that different from DNA base-editing techniques, which typically use Cas9 or other enzymes attached to a CRISPR guide RNA to replace one nucleotide with another. In the case of RNA editing, the enzymes being used in research are predominantly adenosine deaminases acting on RNA (ADARs), which have multiple functions in humans and many other animals, including ensuring that the cell’s own RNA molecules don’t get destroyed by antiviral defenses on the lookout for foreign genetic material.
Endogenous ADARs edit genetic material in the cell by attaching to naturally occurring double-stranded RNAs and switching out A bases with I bases (left panel in the below graphic). Therapeutic RNA editing platforms based on this mechanism fall into one of two categories: either they use engineered enzymes, which generally consist of the editing part of the ADAR enzyme attached to another protein such as Cas13 that boosts specificity, alongside a guide RNA that targets the enzyme to the desired location (middle); or they consist of a guide RNA alone, which recruits endogenous ADAR to edit the target sequence (right).
Read the full story.
