ABOVE: © ISTOCK.COM, kazakovmaksim
In labs around the world, unusual mice scuttle around their cages. Half of each mouse’s tail is coated with fur the shade of burnt toast, while the tip is drained of color. It’s a telltale sign of variations in a gene called kit, yet these mice don’t carry mutations associated with the color mismatch in either of their alleles. Their parents, however, each harbored one mutated copy and have somehow passed a genetic memory on to their litter.
Heritable traits unexplained by genetic sequences are known to occur epigenetically—for example, through the accumulation of molecular caps on DNA that alter gene expression. Typically, researchers have focused on how environmental factors, such as parental stress and malnutrition, alter the epigenetic profile of their offspring. But epigenetic changes might also be guided by the mutations in a parent’s genome, says Didier Stainier, a developmental geneticist at the Max Planck Institute for Heart and Lung Research in Germany.
In a study published November 25 in Science Advances, Stainier and his colleagues report that epigenetic changes of this kind can be inherited in nematode worms and zebrafish, sometimes across multiple generations. The findings “underscore the complexity of biological inheritance beyond the DNA sequence,” says Ana Boskovic, a molecular biologist at the European Molecular Biology Laboratory in Italy who was not involved in the work.
See “Do Epigenetic Changes Influence Evolution?”
Stainier’s lab has been investigating a genetic phenomenon called transcriptional adaptation, where a mutation in one gene boosts the expression of other, so-called adapting genes. The process appears to be dependent on degradation of mutated mRNA, since the effect disappears for mutations that block transcription and in cells where the degradation machinery is silenced. His group has also detected methyl groups crowding the regulatory regions of adapting genes, hinting that the process may involve epigenetic changes.
To uncover whether transcriptional adaptation is inherited, the group bred Caenorhabditis elegans that possessed one functional copy and one mutated copy of a gene called act-5. The researchers had previously shown that this mutated act-5 amplifies expression of another gene, act-3, and they made act-3’s protein product visible by attaching a sequence coding for a red fluorescent protein to the gene.
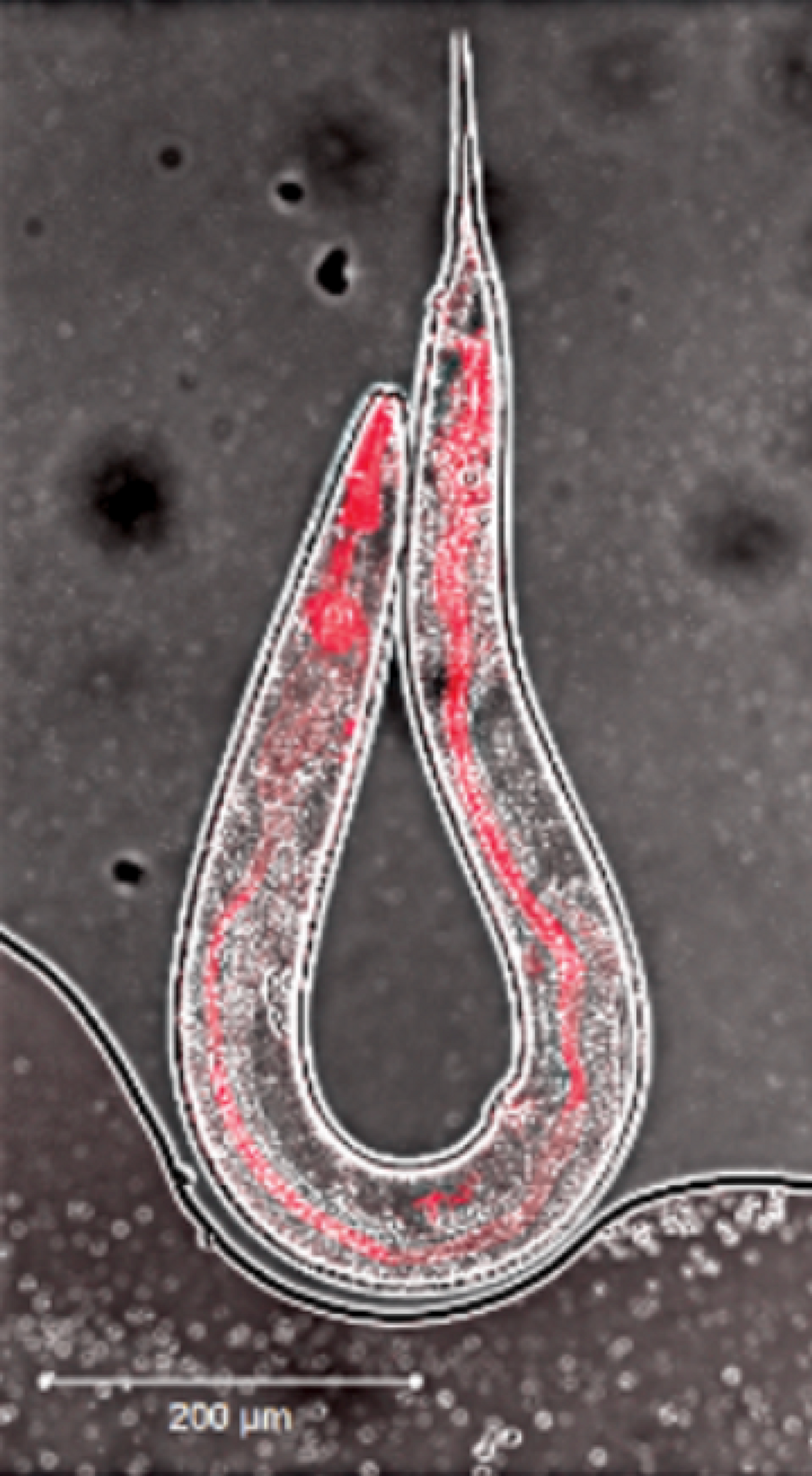
Act-3 is normally only expressed in the pharynx of worms. But in act-5 mutants, act-3 is also activated in the intestine, which lights up red like a piece of tinsel. The intestines of all these mutants’ offspring—including those who didn’t inherit the mutation—lit up, pointing to inherited transcriptional adaptation that boosted act-3 expression. When the researchers continued to breed the mutationless offspring, their guts glowed red for six generations. The signal weakened with each round of breeding, disappearing in the seventh generation.
Wondering whether inherited transcriptional adaptation also occurs in vertebrates, the team turned to zebrafish (Danio rerio) carrying mutations known to upregulate adapting genes. One of these models contained a mutation in a gene called alcama, which enhances expression of the adapting gene alcamb. As with the worms, zebrafish offspring without the mutation showed increased activation of alcamb and a smattering of histone methylation marks on the gene’s regulatory region that weren’t present in descendants of wildtype fish. The researchers achieved the same effect when they injected RNA from a mutated zebrafish’s gametes into an embryo with wildtype parents, demonstrating that the genetic memory stems from RNA present in the germline.
The findings suggest that the “transcriptome depends not only on the mutations you inherited from your parents, but also on those you didn’t inherit,” Stainier tells The Scientist.
Together with the lab’s previous explorations of the mechanism behind transcriptional adaptation, the data suggest that when cells carve up mutated transcripts, scraps of mRNA promote histone methylation of adapted genes, causing heritable changes to gene expression. Stainier’s lab is now trying to understand how pieces of mRNA cause epigenetic changes.
One likely possibility, he says, is that the degraded nucleic acids recruit proteins that remodel chromatin, since small interfering RNAs are known to trigger similar alterations. Or the pieces may interact with antisense transcripts—which normally block translation by binding to mRNA—and thus allow protein synthesis to go ahead uninhibited, says Stainier. “There are a lot of unanswered questions, especially at the mechanistic level,” he says, adding that he hopes the findings will spur research from other groups.
See “Does Human Epigenetic Inheritance Deserve a Closer Look?”
It’s unclear what biological significance transcriptional adaptation might have on offspring, says Miklos Toth, a pharmacologist at Weill Cornell Medical College in New York who did not take part in the study. He compares the process to paramutations, where one allele alters the expression of its genetic partner through heritable epigenetic changes, thought to be controlled by short interfering RNAs. But unlike with paramutations, where offspring inherit obvious features from their parents—for example, maize can inherit purple stems in this way—it is unclear whether transcriptional adaptation has a significant impact on offspring, Toth says.
Stainier admits that he doesn’t know what, if any, advantage inheritance of gene upregulation would have for offspring that don’t inherit their parents’ mutations. When his lab members first uncovered and christened the phenomenon, they found it can compensate for the loss of the mutated gene, he says. But they have since found examples where it can make things worse.
The phenomenon illustrates just how complicated epigenetic mechanisms really are, he adds. “There are so many layers and complexity,” says Stainier. “We are only just scratching the surface.”


